Utilizing Selected Di- and Trinucleotides of siRNA to Predict RNAi Activity
- PMID: 28243313
- PMCID: PMC5294759
- DOI: 10.1155/2017/5043984
Utilizing Selected Di- and Trinucleotides of siRNA to Predict RNAi Activity
Abstract
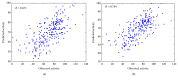
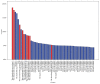
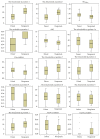
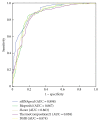
Small interfering RNAs (siRNAs) induce posttranscriptional gene silencing in various organisms. siRNAs targeted to different positions of the same gene show different effectiveness; hence, predicting siRNA activity is a crucial step. In this paper, we developed and evaluated a powerful tool named "siRNApred" with a new mixed feature set to predict siRNA activity. To improve the prediction accuracy, we proposed 2-3NTs as our new features. A Random Forest siRNA activity prediction model was constructed using the feature set selected by our proposed Binary Search Feature Selection (BSFS) algorithm. Experimental data demonstrated that the binding site of the Argonaute protein correlates with siRNA activity. "siRNApred" is effective for selecting active siRNAs, and the prediction results demonstrate that our method can outperform other current siRNA activity prediction methods in terms of prediction accuracy.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


Similar articles
-
Predicting siRNA efficacy based on multiple selective siRNA representations and their combination at score level.Sci Rep. 2017 Mar 20;7:44836. doi: 10.1038/srep44836. Sci Rep. 2017. PMID: 28317874 Free PMC article.
-
The effect of regions flanking target site on siRNA potency.Genomics. 2013 Oct;102(4):215-22. doi: 10.1016/j.ygeno.2013.07.009. Epub 2013 Jul 25. Genomics. 2013. PMID: 23891614
-
Computational models with thermodynamic and composition features improve siRNA design.BMC Bioinformatics. 2006 Feb 12;7:65. doi: 10.1186/1471-2105-7-65. BMC Bioinformatics. 2006. PMID: 16472402 Free PMC article.
-
Design and validation of siRNAs and shRNAs.Curr Opin Mol Ther. 2009 Apr;11(2):156-64. Curr Opin Mol Ther. 2009. PMID: 19330721 Review.
-
Bioinformatics of siRNA design.Methods Mol Biol. 2014;1097:477-90. doi: 10.1007/978-1-62703-709-9_22. Methods Mol Biol. 2014. PMID: 24639173 Review.
Cited by
-
siRNADiscovery: a graph neural network for siRNA efficacy prediction via deep RNA sequence analysis.Brief Bioinform. 2024 Sep 23;25(6):bbae563. doi: 10.1093/bib/bbae563. Brief Bioinform. 2024. PMID: 39503523 Free PMC article.
-
SiRNA silencing efficacy prediction based on a deep architecture.BMC Genomics. 2018 Sep 24;19(Suppl 7):669. doi: 10.1186/s12864-018-5028-8. BMC Genomics. 2018. PMID: 30255786 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

