Chromosome choice for initiation of V-(D)-J recombination is not governed by genomic imprinting
- PMID: 28244489
- PMCID: PMC5788196
- DOI: 10.1038/icb.2017.1
Chromosome choice for initiation of V-(D)-J recombination is not governed by genomic imprinting
Abstract
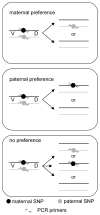
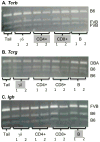
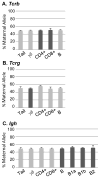
V-(D)-J recombination generates the antigen receptor diversity necessary for immune cell function, while allelic exclusion ensures that each cell expresses a single antigen receptor. V-(D)-J recombination of the Ig, Tcrb, Tcrg and Tcrd antigen receptor genes is ordered and sequential so that only one allele generates a productive rearrangement. The mechanism controlling sequential rearrangement of antigen receptor genes, in particular how only one allele is selected to initiate recombination while at least temporarily leaving the other intact, remains unresolved. Genomic imprinting, a widespread phenomenon wherein maternal or paternal allele inheritance determines allele activity, could represent a regulatory mechanism for controlling sequential V-(D)-J rearrangement. We used strain-specific single-nucleotide polymorphisms within antigen receptor genes to determine if maternal vs paternal inheritance could underlie chromosomal choice for the initiation of recombination. We found no parental chromosomal bias in the initiation of V-(D)-J recombination in T or B cells, eliminating genomic imprinting as a potential regulator for this tightly regulated process.
Conflict of interest statement
The authors declare no conflict of interest.
The authors have no competing financial interests to report.
Figures
References
-
- Malissen M, Trucy J, Jouvin-Marche E, Cazenave P, Scollay R, Malissen B. Regulation of TCR alpha and beta gene allelic exclusion during T-cell development. Immunol Today. 1992;13:315–22. - PubMed
-
- Outters P, Jaeger S, Zaarour N, Ferrier P. Long-range control of V(D)J recombination and allelic exclusion: modelling views. Adv Immunol. 2015;128:363–413. - PubMed
-
- Jackson A, Krangel M. Turning T-cell receptor beta recombination on and off: more questions than answers. Immunol Rev. 2006;209:129–41. - PubMed
-
- Khor B, Sleckman B. Allelic exclusion at the TCRbeta locus. Curr Opin Immunol. 2002;14:230–34. - PubMed
-
- Michie A, Zuniga-Pflucker J. Regulation of thymocyte differentiation: pre-TCR signals and beta-selection. Semin Immunol. 2002;14:3311–323. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

