RNA-protein binding motifs mining with a new hybrid deep learning based cross-domain knowledge integration approach
- PMID: 28245811
- PMCID: PMC5331642
- DOI: 10.1186/s12859-017-1561-8
RNA-protein binding motifs mining with a new hybrid deep learning based cross-domain knowledge integration approach
Abstract
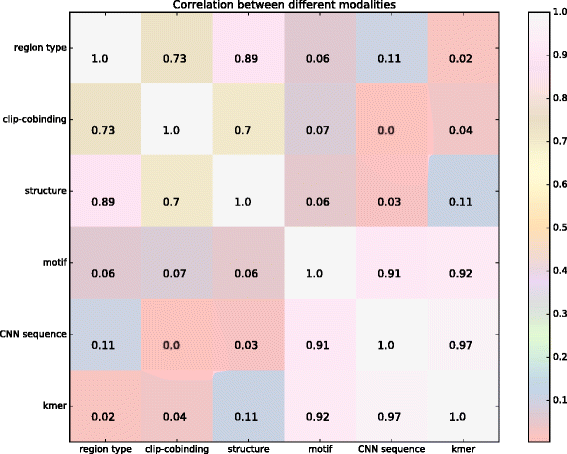
Background: RNAs play key roles in cells through the interactions with proteins known as the RNA-binding proteins (RBP) and their binding motifs enable crucial understanding of the post-transcriptional regulation of RNAs. How the RBPs correctly recognize the target RNAs and why they bind specific positions is still far from clear. Machine learning-based algorithms are widely acknowledged to be capable of speeding up this process. Although many automatic tools have been developed to predict the RNA-protein binding sites from the rapidly growing multi-resource data, e.g. sequence, structure, their domain specific features and formats have posed significant computational challenges. One of current difficulties is that the cross-source shared common knowledge is at a higher abstraction level beyond the observed data, resulting in a low efficiency of direct integration of observed data across domains. The other difficulty is how to interpret the prediction results. Existing approaches tend to terminate after outputting the potential discrete binding sites on the sequences, but how to assemble them into the meaningful binding motifs is a topic worth of further investigation.
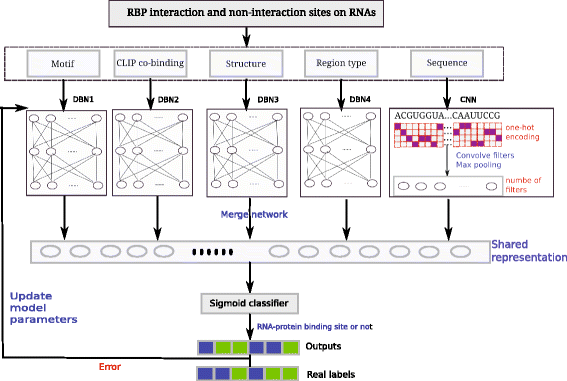
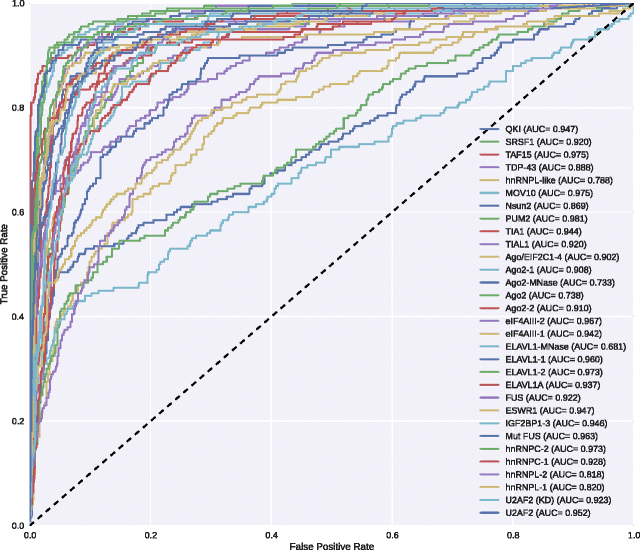
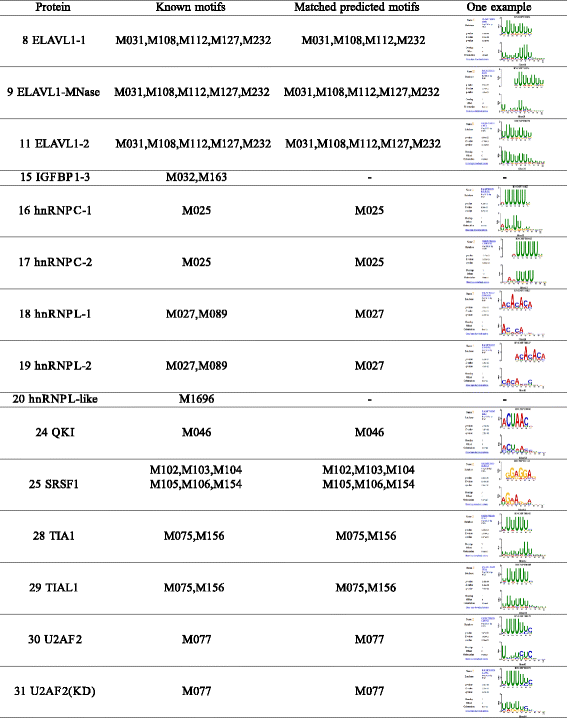
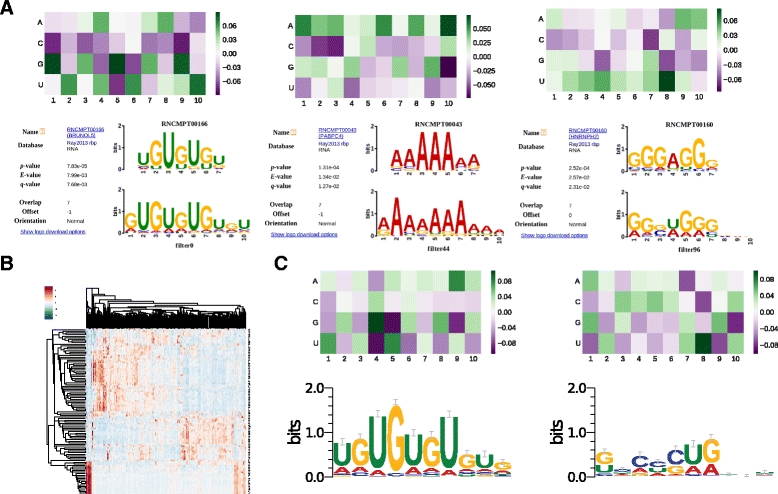
Results: In viewing of these challenges, we propose a deep learning-based framework (iDeep) by using a novel hybrid convolutional neural network and deep belief network to predict the RBP interaction sites and motifs on RNAs. This new protocol is featured by transforming the original observed data into a high-level abstraction feature space using multiple layers of learning blocks, where the shared representations across different domains are integrated. To validate our iDeep method, we performed experiments on 31 large-scale CLIP-seq datasets, and our results show that by integrating multiple sources of data, the average AUC can be improved by 8% compared to the best single-source-based predictor; and through cross-domain knowledge integration at an abstraction level, it outperforms the state-of-the-art predictors by 6%. Besides the overall enhanced prediction performance, the convolutional neural network module embedded in iDeep is also able to automatically capture the interpretable binding motifs for RBPs. Large-scale experiments demonstrate that these mined binding motifs agree well with the experimentally verified results, suggesting iDeep is a promising approach in the real-world applications.
Conclusion: The iDeep framework not only can achieve promising performance than the state-of-the-art predictors, but also easily capture interpretable binding motifs. iDeep is available at http://www.csbio.sjtu.edu.cn/bioinf/iDeep.
Keywords: CLIP-seq; Convolutional neural network; Deep belief network; Multimodal deep learning; RNA-binding protein.
Figures

Similar articles
-
Prediction of RNA-protein sequence and structure binding preferences using deep convolutional and recurrent neural networks.BMC Genomics. 2018 Jul 3;19(1):511. doi: 10.1186/s12864-018-4889-1. BMC Genomics. 2018. PMID: 29970003 Free PMC article.
-
RNA-binding protein recognition based on multi-view deep feature and multi-label learning.Brief Bioinform. 2021 May 20;22(3):bbaa174. doi: 10.1093/bib/bbaa174. Brief Bioinform. 2021. PMID: 32808039
-
Predicting RNA-protein binding sites and motifs through combining local and global deep convolutional neural networks.Bioinformatics. 2018 Oct 15;34(20):3427-3436. doi: 10.1093/bioinformatics/bty364. Bioinformatics. 2018. PMID: 29722865
-
Comprehensive review and assessment of computational methods for predicting RNA post-transcriptional modification sites from RNA sequences.Brief Bioinform. 2020 Sep 25;21(5):1676-1696. doi: 10.1093/bib/bbz112. Brief Bioinform. 2020. PMID: 31714956 Review.
-
Computational analysis of CLIP-seq data.Methods. 2017 Apr 15;118-119:60-72. doi: 10.1016/j.ymeth.2017.02.006. Epub 2017 Feb 22. Methods. 2017. PMID: 28254606 Review.
Cited by
-
Prediction of mRNA subcellular localization using deep recurrent neural networks.Bioinformatics. 2019 Jul 15;35(14):i333-i342. doi: 10.1093/bioinformatics/btz337. Bioinformatics. 2019. PMID: 31510698 Free PMC article.
-
Large-scale prediction of protein ubiquitination sites using a multimodal deep architecture.BMC Syst Biol. 2018 Nov 22;12(Suppl 6):109. doi: 10.1186/s12918-018-0628-0. BMC Syst Biol. 2018. PMID: 30463553 Free PMC article.
-
RBPSpot: Learning on appropriate contextual information for RBP binding sites discovery.iScience. 2021 Oct 30;24(12):103381. doi: 10.1016/j.isci.2021.103381. eCollection 2021 Dec 17. iScience. 2021. PMID: 34841226 Free PMC article.
-
Assessing deep learning methods in cis-regulatory motif finding based on genomic sequencing data.Brief Bioinform. 2022 Jan 17;23(1):bbab374. doi: 10.1093/bib/bbab374. Brief Bioinform. 2022. PMID: 34607350 Free PMC article.
-
A self-attention model for inferring cooperativity between regulatory features.Nucleic Acids Res. 2021 Jul 21;49(13):e77. doi: 10.1093/nar/gkab349. Nucleic Acids Res. 2021. PMID: 33950192 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

