A streamlined method for the design and cloning of shRNAs into an optimized Dox-inducible lentiviral vector
- PMID: 28245848
- PMCID: PMC5331646
- DOI: 10.1186/s12896-017-0341-x
A streamlined method for the design and cloning of shRNAs into an optimized Dox-inducible lentiviral vector
Abstract
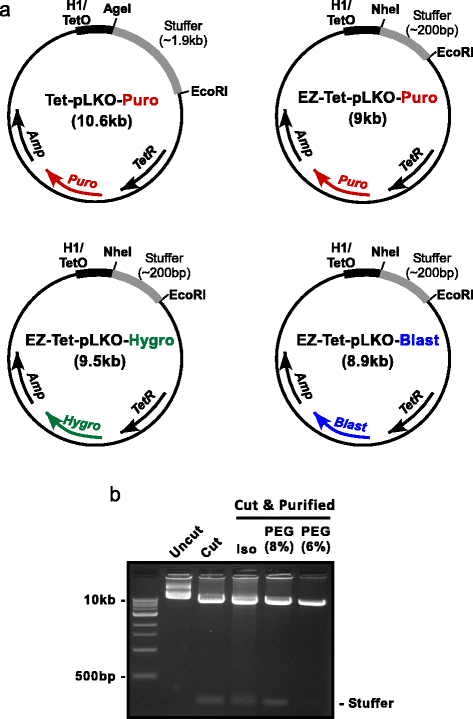
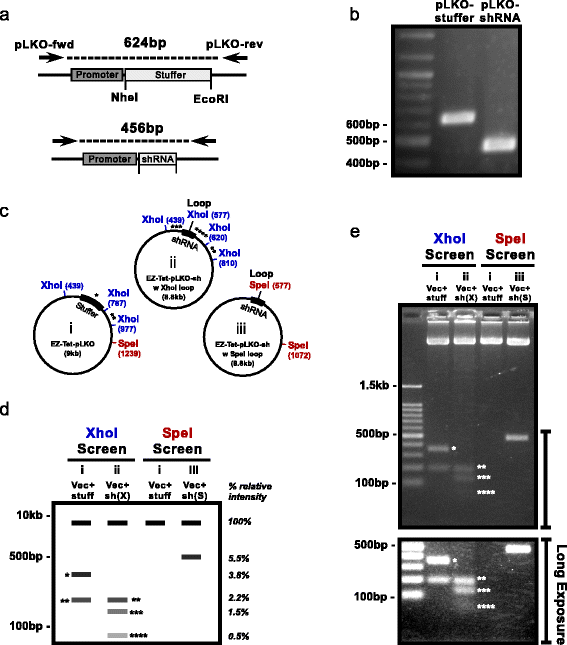
Background: Short hairpin RNA (shRNA) is an established and effective tool for stable knock down of gene expression. Lentiviral vectors can be used to deliver shRNAs, thereby providing the ability to infect most mammalian cell types with high efficiency, regardless of proliferation state. Furthermore, the use of inducible promoters to drive shRNA expression allows for more thorough investigations into the specific timing of gene function in a variety of cellular processes. Moreover, inducible knockdown allows the investigation of genes that would be lethal or otherwise poorly tolerated if constitutively knocked down. Lentiviral inducible shRNA vectors are readily available, but unfortunately the process of cloning, screening, and testing shRNAs can be time-consuming and expensive. Therefore, we sought to refine a popular vector (Tet-pLKO-Puro) and streamline the cloning process with efficient protocols so that researchers can more efficiently utilize this powerful tool. METHODS: First, we modified the Tet-pLKO-Puro vector to make it easy ("EZ") for molecular cloning (EZ-Tet-pLKO-Puro). Our primary modification was to shrink the stuffer region, which allows vector purification via polyethylene glycol precipitation thereby avoiding the need to purify DNA through agarose. In addition, we generated EZ-Tet-pLKO vectors with hygromycin or blasticidin resistance to provide greater flexibility in cell line engineering. Furthermore, we provide a detailed guide for utilizing these vectors, including shRNA design strategy and simplified screening methods.
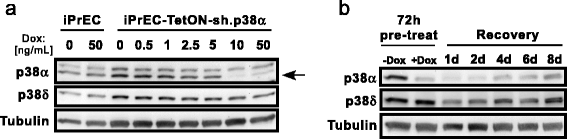
Results: Notably, we emphasize the importance of loop sequence design and demonstrate that the addition of a single mismatch in the loop stem can greatly improve shRNA efficiency. Lastly, we display the robustness of the system with a doxycycline titration and recovery time course and provide a cost/benefit analysis comparing our system with purchasing pre-designed shRNA vectors.
Conclusions: Our aim was twofold: first, to take a very useful shRNA vector and make it more amenable for molecular cloning and, secondly, to provide a streamlined protocol and rationale for cost-effective design, cloning, and screening of shRNAs. With this knowledge, anyone can take advantage of this powerful tool to inducibly knockdown any gene of their choosing.
Keywords: Inducible; Lentivirus; pLKO; shRNA.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

