A mammalian homolog of the zebrafish transmembrane protein 2 (TMEM2) is the long-sought-after cell-surface hyaluronidase
- PMID: 28246172
- PMCID: PMC5418033
- DOI: 10.1074/jbc.M116.770149
A mammalian homolog of the zebrafish transmembrane protein 2 (TMEM2) is the long-sought-after cell-surface hyaluronidase
Abstract
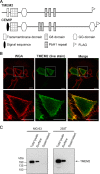
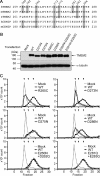
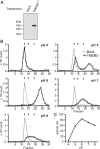
Hyaluronan (HA) is an extremely large polysaccharide (glycosaminoglycan) involved in many cellular functions. HA catabolism is thought to involve the initial cleavage of extracellular high-molecular-weight (HMW) HA into intermediate-size HA by an extracellular or cell-surface hyaluronidase, internalization of intermediate-size HA, and complete degradation into monosaccharides in lysosomes. Despite considerable research, the identity of the hyaluronidase responsible for the initial HA cleavage in the extracellular space remains elusive. HYAL1 and HYAL2 have properties more consistent with lysosomal hyaluronidases, whereas CEMIP/KIAA1199, a recently identified HA-binding molecule that has HA-degrading activity, requires the participation of the clathrin-coated pit pathway of live cells for HA degradation. Here we show that transmembrane protein 2 (TMEM2), a mammalian homolog of a protein playing a role in zebrafish endocardial cushion development, is a cell-surface hyaluronidase. Live immunostaining and surface biotinylation assays confirmed that mouse TMEM2 is expressed on the cell surface in a type II transmembrane topology. TMEM2 degraded HMW-HA into ∼5-kDa fragments but did not cleave chondroitin sulfate or dermatan sulfate, indicating its specificity to HA. The hyaluronidase activity of TMEM2 was Ca2+-dependent; the enzyme's pH optimum is around 6-7, and unlike CEMIP/KIAA1199, TMEM2 does not require the participation of live cells for its hyaluronidase activity. Moreover, TMEM2-expressing cells could eliminate HA immobilized on a glass surface in a contact-dependent manner. Together, these data suggest that TMEM2 is the long-sought-after hyaluronidase that cleaves extracellular HMW-HA into intermediate-size fragments before internalization and degradation in the lysosome.
Keywords: CEMIP/KIAA1199; TMEM2; cell surface; glycosaminoglycan; hyaluronan; hyaluronidase; membrane function.
© 2017 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures



Similar articles
-
The cell surface hyaluronidase TMEM2 is essential for systemic hyaluronan catabolism and turnover.J Biol Chem. 2021 Nov;297(5):101281. doi: 10.1016/j.jbc.2021.101281. Epub 2021 Oct 6. J Biol Chem. 2021. PMID: 34624311 Free PMC article.
-
TMEM2 is a bona fide hyaluronidase possessing intrinsic catalytic activity.J Biol Chem. 2023 Sep;299(9):105120. doi: 10.1016/j.jbc.2023.105120. Epub 2023 Jul 30. J Biol Chem. 2023. PMID: 37527776 Free PMC article.
-
The role and regulation of TMEM2 (transmembrane protein 2) in HYBID (hyaluronan (HA)-binding protein involved in HA depolymerization/ KIAA1199/CEMIP)-mediated HA depolymerization in human skin fibroblasts.Biochem Biophys Res Commun. 2018 Oct 20;505(1):74-80. doi: 10.1016/j.bbrc.2018.09.097. Epub 2018 Sep 18. Biochem Biophys Res Commun. 2018. PMID: 30241936
-
TMEM2: A missing link in hyaluronan catabolism identified?Matrix Biol. 2019 May;78-79:139-146. doi: 10.1016/j.matbio.2018.03.020. Epub 2018 Mar 27. Matrix Biol. 2019. PMID: 29601864 Free PMC article. Review.
-
New molecules indispensable for hyaluronan degradation, HYBID (CEMIP/KIAA1199) and TMEM2 (CEMIP2): Differential roles in physiological and pathological non-neoplastic conditions.Proc Jpn Acad Ser B Phys Biol Sci. 2025;101(6):317-338. doi: 10.2183/pjab.101.021. Proc Jpn Acad Ser B Phys Biol Sci. 2025. PMID: 40500182 Review.
Cited by
-
In-silico analysis of TMEM2 as a pancreatic adenocarcinoma and cancer-associated fibroblast biomarker, and functional characterization of NSC777201, for targeted drug development.Am J Cancer Res. 2024 Jun 15;14(6):3010-3035. doi: 10.62347/CHXD6134. eCollection 2024. Am J Cancer Res. 2024. PMID: 39005682 Free PMC article.
-
Key Matrix Remodeling Enzymes: Functions and Targeting in Cancer.Cancers (Basel). 2021 Mar 22;13(6):1441. doi: 10.3390/cancers13061441. Cancers (Basel). 2021. PMID: 33809973 Free PMC article. Review.
-
Possible Repositioning of an Oral Anti-Osteoporotic Drug, Ipriflavone, for Treatment of Inflammatory Arthritis via Inhibitory Activity of KIAA1199, a Novel Potent Hyaluronidase.Int J Mol Sci. 2022 Apr 7;23(8):4089. doi: 10.3390/ijms23084089. Int J Mol Sci. 2022. PMID: 35456905 Free PMC article.
-
Tmem2 restricts atrioventricular canal differentiation by regulating degradation of hyaluronic acid.Dev Dyn. 2019 Dec;248(12):1195-1210. doi: 10.1002/dvdy.106. Epub 2019 Sep 10. Dev Dyn. 2019. PMID: 31444829 Free PMC article.
-
The Degradation of Hyaluronan in the Skin.Biomolecules. 2022 Feb 3;12(2):251. doi: 10.3390/biom12020251. Biomolecules. 2022. PMID: 35204753 Free PMC article.
References
-
- Laurent T. C., and Fraser J. R. (1992) Hyaluronan. FASEB J. 6, 2397–2404 - PubMed
-
- Fraser J. R., and Laurent T. C. (1989) Turnover and metabolism of hyaluronan. Ciba Found. Symp. 143, 41–53 - PubMed
-
- Stern R. (2003) Devising a pathway for hyaluronan catabolism: are we there yet? Glycobiology 13, 105R–115R - PubMed
-
- Stern R., Kogan G., Jedrzejas M. J., and Soltés L. (2007) The many ways to cleave hyaluronan. Biotechnol. Adv. 25, 537–557 - PubMed
-
- Chanmee T., Ontong P., and Itano N. (2016) Hyaluronan: A modulator of the tumor microenvironment. Cancer Lett. 375, 20–30 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

