Stilbene epoxidation and detoxification in a Photorhabdus luminescens-nematode symbiosis
- PMID: 28246174
- PMCID: PMC5399116
- DOI: 10.1074/jbc.M116.762542
Stilbene epoxidation and detoxification in a Photorhabdus luminescens-nematode symbiosis
Abstract
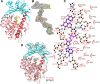
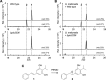
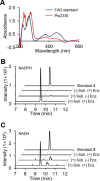
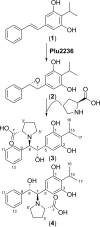
Members of the gammaproteobacterial Photorhabdus genus share mutualistic relationships with Heterorhabditis nematodes, and the pairs infect a wide swath of insect larvae. Photorhabdus species produce a family of stilbenes, with two major components being 3,5-dihydroxy-4-isopropyl-trans-stilbene (compound 1) and its stilbene epoxide (compound 2). This family of molecules harbors antimicrobial and immunosuppressive activities, and its pathway is responsible for producing a nematode "food signal" involved in nematode development. However, stilbene epoxidation biosynthesis and its biological roles remain unknown. Here, we identified an orphan protein (Plu2236) from Photorhabdus luminescens that catalyzes stilbene epoxidation. Structural, mutational, and biochemical analyses confirmed the enzyme adopts a fold common to FAD-dependent monooxygenases, contains a tightly bound FAD prosthetic group, and is required for the stereoselective epoxidation of compounds 1 and 2. The epoxidase gene was dispensable in a nematode-infective juvenile recovery assay, indicating the oxidized compound is not required for the food signal. The epoxide exhibited reduced cytotoxicity toward its producer, suggesting this may be a natural route for intracellular detoxification. In an insect infection model, we also observed two stilbene-derived metabolites that were dependent on the epoxidase. NMR, computational, and chemical degradation studies established their structures as new stilbene-l-proline conjugates, prolbenes A (compound 3) and B (compound 4). The prolbenes lacked immunosuppressive and antimicrobial activities compared with their stilbene substrates, suggesting a metabolite attenuation mechanism in the animal model. Collectively, our studies provide a structural view for stereoselective stilbene epoxidation and functionalization in an invertebrate animal infection model and provide new insights into stilbene cellular detoxification.
Keywords: Gram-negative bacteria; crystal structure; natural product; secondary metabolism; symbiosis.
© 2017 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures




Similar articles
-
Iso-propyl stilbene: a life cycle signal?Microbiology (Reading). 2019 May;165(5):516-526. doi: 10.1099/mic.0.000790. Epub 2019 Mar 18. Microbiology (Reading). 2019. PMID: 30882293
-
HdfR is a regulator in Photorhabdus luminescens that modulates metabolism and symbiosis with the nematode Heterorhabditis.Environ Microbiol. 2012 Apr;14(4):953-66. doi: 10.1111/j.1462-2920.2011.02669.x. Epub 2011 Dec 12. Environ Microbiol. 2012. PMID: 22151606
-
A metabolic switch is involved in lifestyle decisions in Photorhabdus luminescens.Mol Microbiol. 2010 Sep;77(6):1394-405. doi: 10.1111/j.1365-2958.2010.07300.x. Epub 2010 Jul 30. Mol Microbiol. 2010. PMID: 20662779
-
Photorhabdus: a tale of contrasting interactions.Microbiology (Reading). 2020 Apr;166(4):335-348. doi: 10.1099/mic.0.000907. Epub 2020 Mar 24. Microbiology (Reading). 2020. PMID: 32209172 Review.
-
Photorhabdus and a host of hosts.Annu Rev Microbiol. 2009;63:557-74. doi: 10.1146/annurev.micro.091208.073507. Annu Rev Microbiol. 2009. PMID: 19575559 Review.
Cited by
-
The Isopropylstilbene Precursor Cinnamic Acid Inhibits Anthraquinone Pigment Production by Targeting AntI.J Am Chem Soc. 2025 Jun 18;147(24):20246-20250. doi: 10.1021/jacs.5c07388. Epub 2025 Jun 5. J Am Chem Soc. 2025. PMID: 40474393 Free PMC article.
-
Prokaryotic Solute/Sodium Symporters: Versatile Functions and Mechanisms of a Transporter Family.Int J Mol Sci. 2021 Feb 13;22(4):1880. doi: 10.3390/ijms22041880. Int J Mol Sci. 2021. PMID: 33668649 Free PMC article. Review.
-
A Conserved Nonribosomal Peptide Synthetase in Xenorhabdus bovienii Produces Citrulline-Functionalized Lipopeptides.J Nat Prod. 2021 Oct 22;84(10):2692-2699. doi: 10.1021/acs.jnatprod.1c00573. Epub 2021 Sep 28. J Nat Prod. 2021. PMID: 34581573 Free PMC article.
-
Bacterial Autoimmune Drug Metabolism Transforms an Immunomodulator into Structurally and Functionally Divergent Antibiotics.Angew Chem Int Ed Engl. 2020 May 11;59(20):7871-7880. doi: 10.1002/anie.201916204. Epub 2020 Mar 17. Angew Chem Int Ed Engl. 2020. PMID: 32097515 Free PMC article.
-
Role of Proline in Pathogen and Host Interactions.Antioxid Redox Signal. 2019 Feb 1;30(4):683-709. doi: 10.1089/ars.2017.7335. Epub 2018 Feb 2. Antioxid Redox Signal. 2019. PMID: 29241353 Free PMC article. Review.
References
-
- Clarke D. J. (2008) Photorhabdus: a model for the analysis of pathogenicity and mutualism. Cell. Microbiol. 10, 2159–2167 - PubMed
-
- Clarke D. J. (2014) The genetic basis of the symbiosis between Photorhabdus and its invertebrate hosts. Adv. Appl. Microbiol. 88, 1–29 - PubMed
-
- Waterfield N. R., Ciche T., and Clarke D. (2009) Photorhabdus and a host of hosts. Annu. Rev. Microbiol. 63, 557–574 - PubMed
-
- Gerrard J., Waterfield N., Vohra R., and ffrench-Constant R. (2004) Human infection with Photorhabdus asymbiotica: an emerging bacterial pathogen. Microbes Infect. 6, 229–237 - PubMed
-
- Bode H. B. (2009) Entomopathogenic bacteria as a source of secondary metabolites. Curr. Opin. Chem. Biol. 13, 224–230 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

