Synthetic Botany
- PMID: 28246181
- PMCID: PMC5495061
- DOI: 10.1101/cshperspect.a023887
Synthetic Botany
Abstract
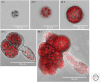
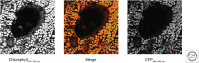
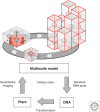
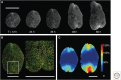
Plants are attractive platforms for synthetic biology and metabolic engineering. Plants' modular and plastic body plans, capacity for photosynthesis, extensive secondary metabolism, and agronomic systems for large-scale production make them ideal targets for genetic reprogramming. However, efforts in this area have been constrained by slow growth, long life cycles, the requirement for specialized facilities, a paucity of efficient tools for genetic manipulation, and the complexity of multicellularity. There is a need for better experimental and theoretical frameworks to understand the way genetic networks, cellular populations, and tissue-wide physical processes interact at different scales. We highlight new approaches to the DNA-based manipulation of plants and the use of advanced quantitative imaging techniques in simple plant models such as Marchantia polymorpha. These offer the prospects of improved understanding of plant dynamics and new approaches to rational engineering of plant traits.
Copyright © 2017 Cold Spring Harbor Laboratory Press; all rights reserved.
Figures

References
-
- Adachi T, Takase H, Tomizawa K. 2007. Introduction of a 50 kbp DNA fragment into the plastid genome. Biosci Biotechnol Biochem 71: 2266–2273. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
