Rapid and efficient generation of oligodendrocytes from human induced pluripotent stem cells using transcription factors
- PMID: 28246330
- PMCID: PMC5358375
- DOI: 10.1073/pnas.1614412114
Rapid and efficient generation of oligodendrocytes from human induced pluripotent stem cells using transcription factors
Abstract
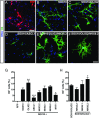
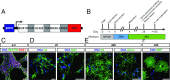
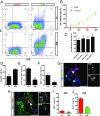
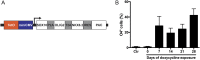
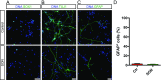
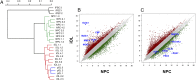
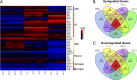
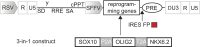
Rapid and efficient protocols to generate oligodendrocytes (OL) from human induced pluripotent stem cells (iPSC) are currently lacking, but may be a key technology to understand the biology of myelin diseases and to develop treatments for such disorders. Here, we demonstrate that the induction of three transcription factors (SOX10, OLIG2, NKX6.2) in iPSC-derived neural progenitor cells is sufficient to rapidly generate O4+ OL with an efficiency of up to 70% in 28 d and a global gene-expression profile comparable to primary human OL. We further demonstrate that iPSC-derived OL disperse and myelinate the CNS of Mbpshi/shiRag-/- mice during development and after demyelination, are suitable for in vitro myelination assays, disease modeling, and screening of pharmacological compounds potentially promoting oligodendroglial differentiation. Thus, the strategy presented here to generate OL from iPSC may facilitate the studying of human myelin diseases and the development of high-throughput screening platforms for drug discovery.
Keywords: disease modeling; forward patterning; human iPSC; myelination; oligodendrocytes.
Conflict of interest statement
Conflict of interest statement: M.E. and T.K. have a pending patent application for the oligodendroglial differentiation protocol.
Figures
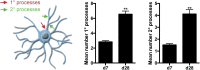


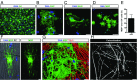



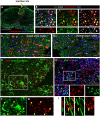
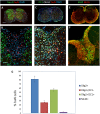
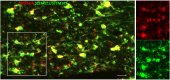
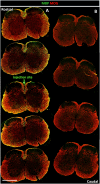
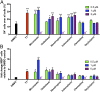
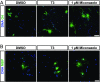
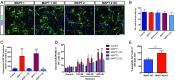
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

