Breaking up and making up: The secret life of the vacuolar H+ -ATPase
- PMID: 28247968
- PMCID: PMC5405435
- DOI: 10.1002/pro.3147
Breaking up and making up: The secret life of the vacuolar H+ -ATPase
Abstract
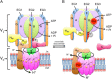
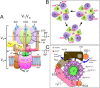
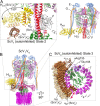
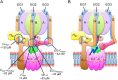
The vacuolar ATPase (V-ATPase; V1 Vo -ATPase) is a large multisubunit proton pump found in the endomembrane system of all eukaryotic cells where it acidifies the lumen of subcellular organelles including lysosomes, endosomes, the Golgi apparatus, and clathrin-coated vesicles. V-ATPase function is essential for pH and ion homeostasis, protein trafficking, endocytosis, mechanistic target of rapamycin (mTOR), and Notch signaling, as well as hormone secretion and neurotransmitter release. V-ATPase can also be found in the plasma membrane of polarized animal cells where its proton pumping function is involved in bone remodeling, urine acidification, and sperm maturation. Aberrant (hypo or hyper) activity has been associated with numerous human diseases and the V-ATPase has therefore been recognized as a potential drug target. Recent progress with moderate to high-resolution structure determination by cryo electron microscopy and X-ray crystallography together with sophisticated single-molecule and biochemical experiments have provided a detailed picture of the structure and unique mode of regulation of the V-ATPase. This review summarizes the recent advances, focusing on the structural and biophysical aspects of the field.
Keywords: V-ATPase; V1Vo-ATPase; X-ray crystallography; cryo electron microscopy; protein structure; protein-protein interactions; reversible disassembly; rotary catalysis; rotary motor enzyme; vacuolar ATPase.
© 2017 The Protein Society.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

