Comparing Simplification Strategies for the Skeletal Muscle Proteome
- PMID: 28248220
- PMCID: PMC5217366
- DOI: 10.3390/proteomes4010010
Comparing Simplification Strategies for the Skeletal Muscle Proteome
Abstract
Skeletal muscle is a complex tissue that is dominated by the presence of a few abundant proteins. This wide dynamic range can mask the presence of lower abundance proteins, which can be a confounding factor in large-scale proteomic experiments. In this study, we have investigated a number of pre-fractionation methods, at both the protein and peptide level, for the characterization of the skeletal muscle proteome. The analyses revealed that the use of OFFGEL isoelectric focusing yielded the largest number of protein identifications (>750) compared to alternative gel-based and protein equalization strategies. Further, OFFGEL led to a substantial enrichment of a different sub-population of the proteome. Filter-aided sample preparation (FASP), coupled to peptide-level OFFGEL provided more confidence in the results due to a substantial increase in the number of peptides assigned to each protein. The findings presented here support the use of a multiplexed approach to proteome characterization of skeletal muscle, which has a recognized imbalance in the dynamic range of its protein complement.
Keywords: FASP; OFFGEL; ProteoMiner; protein equalization; proteome simplification; skeletal muscle.
Figures
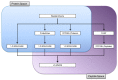

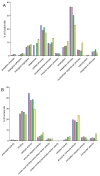

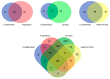
Similar articles
-
Low-molecular-weight color pI markers to monitor on-line the peptide focusing process in OFFGEL fractionation.Electrophoresis. 2017 Aug;38(16):2034-2041. doi: 10.1002/elps.201700075. Epub 2017 Jul 26. Electrophoresis. 2017. PMID: 28672066
-
Improvements in proteomic metrics of low abundance proteins through proteome equalization using ProteoMiner prior to MudPIT.J Proteome Res. 2011 Aug 5;10(8):3690-700. doi: 10.1021/pr200304u. Epub 2011 Jun 24. J Proteome Res. 2011. PMID: 21702434 Free PMC article.
-
Use of liquid isoelectric focusing (OFFGEL) on the discovery of meat tenderness biomarkers.J Proteomics. 2018 Jul 15;183:25-33. doi: 10.1016/j.jprot.2018.05.005. Epub 2018 May 9. J Proteomics. 2018. PMID: 29751105
-
A review on preparative and semi-preparative offgel electrophoresis for multidimensional protein/peptide assessment.Anal Chim Acta. 2014 Jul 11;836:1-17. doi: 10.1016/j.aca.2014.04.053. Epub 2014 May 1. Anal Chim Acta. 2014. PMID: 24974865 Review.
-
Concepts and strategies of soybean seed proteomics using the shotgun proteomics approach.Expert Rev Proteomics. 2019 Sep;16(9):795-804. doi: 10.1080/14789450.2019.1654860. Epub 2019 Aug 13. Expert Rev Proteomics. 2019. PMID: 31398080 Review.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources

