A three-microRNA signature identifies two subtypes of glioblastoma patients with different clinical outcomes
- PMID: 28248456
- PMCID: PMC5579331
- DOI: 10.1002/1878-0261.12047
A three-microRNA signature identifies two subtypes of glioblastoma patients with different clinical outcomes
Abstract
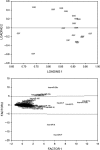
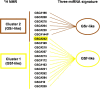
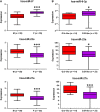
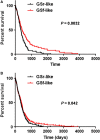
Glioblastoma multiforme (GBM) is the most common and malignant primary brain tumor in adults, characterized by aggressive growth, limited response to therapy, and inexorable recurrence. Because of the extremely unfavorable prognosis of GBM, it is important to develop more effective diagnostic and therapeutic strategies based on biologically and clinically relevant patient stratification systems. Analyzing a collection of patient-derived GBM stem-like cells (GSCs) by gene expression profiling, nuclear magnetic resonance spectroscopy, and signal transduction pathway activation, we identified two GSC clusters characterized by different clinical features. Due to the widely documented role played by microRNAs (miRNAs) in the tumorigenesis process, in this study we explored whether these two GBM patient subtypes could also be discriminated by different miRNA signatures. Global miRNA expression pattern was analyzed by oblique principal component analysis and principal component analysis. By a combined inferential strategy on PCA results, we identified a reduced set of three miRNAs - miR-23a, miR-27a, and miR-9* (miR-9-3p) - able to discriminate the proneural- and mesenchymal-like GSC phenotypes as well as mesenchymal and proneural subtypes of primary GBM included in The Cancer Genome Atlas (TCGA) data set. Kaplan-Meier analysis showed a significant correlation between the selected miRNAs and overall survival in 429 GBM specimens from TCGA-identifying patients who had an unfavorable outcome. The survival prognostic capability of the three-miRNA signatures could have important implications for the understanding of the biology of GBM subtypes and could be useful in patient stratification to facilitate interpretation of results from clinical trials.
Keywords: glioblastoma; glioblastoma stem-like cells; microRNAs; patient stratification.
© 2017 The Authors. Published by FEBS Press and John Wiley & Sons Ltd.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

