Diversification of Orientia tsutsugamushi genotypes by intragenic recombination and their potential expansion in endemic areas
- PMID: 28248956
- PMCID: PMC5348041
- DOI: 10.1371/journal.pntd.0005408
Diversification of Orientia tsutsugamushi genotypes by intragenic recombination and their potential expansion in endemic areas
Abstract
Background: Scrub typhus is a mite-borne febrile disease caused by O. tsutsugamushi infection. Recently, emergence of scrub typhus has attracted considerable attention in several endemic countries in Asia and the western Pacific. In addition, the antigenic diversity of the intracellular pathogen has been a serious obstacle for developing effective diagnostics and vaccine.
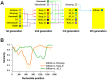
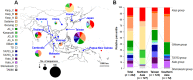
Methodology/principal findings: To understand the evolutionary pathway of genotypic diversification of O. tsutsugamushi and the environmental factors associated with the epidemiological features of scrub typhus, we analyzed sequence data, including spatiotemporal information, of the tsa56 gene encoding a major outer membrane protein responsible for antigenic variation. A total of 324 tsa56 sequences covering more than 85% of its open reading frame were analyzed and classified into 17 genotypes based on phylogenetic relationship. Extensive sequence analysis of tsa56 genes using diverse informatics tools revealed multiple intragenic recombination events, as well as a substantially higher mutation rate than other house-keeping genes. This suggests that genetic diversification occurred via frequent point mutations and subsequent genetic recombination. Interestingly, more diverse bacterial genotypes and dominant vector species prevail in Taiwan compared to other endemic regions. Furthermore, the co-presence of identical and sub-identical clones of tsa56 gene in geographically distant areas implies potential spread of O. tsutsugamushi genotypes.
Conclusions/significance: Fluctuation and diversification of vector species harboring O. tsutsugamushi in local endemic areas may facilitate genetic recombination among diverse genotypes. Therefore, careful monitoring of dominant vector species, as well as the prevalence of O. tsutsugamushi genotypes may be advisable to enable proper anticipation of epidemiological changes of scrub typhus.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





References
-
- Kelly DJ, Fuerst PA, Ching WM, Richards AL (2009) Scrub typhus: the geographic distribution of phenotypic and genotypic variants of Orientia tsutsugamushi. Clin Infect Dis 48 Suppl 3: S203–230. - PubMed
-
- Kawamura A, Tanaka H, Tamura A (1995) Tsutsugamushi disease. Tokyo, Japan: University of Tokyo press.
-
- Ghorbani RP, Ghorbani AJ, Jain MK, Walker DH (1997) A case of scrub typhus probably acquired in Africa. Clin Infect Dis 25: 1473–1474. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

