Genome-wide transcriptomics of aging in the rotifer Brachionus manjavacas, an emerging model system
- PMID: 28249563
- PMCID: PMC5333405
- DOI: 10.1186/s12864-017-3540-x
Genome-wide transcriptomics of aging in the rotifer Brachionus manjavacas, an emerging model system
Abstract
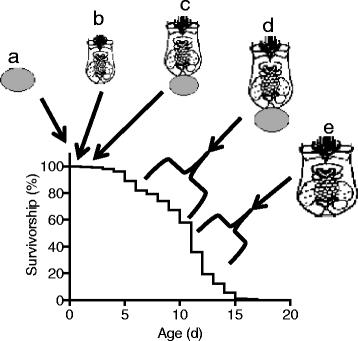
Background: Understanding gene expression changes over lifespan in diverse animal species will lead to insights to conserved processes in the biology of aging and allow development of interventions to improve health. Rotifers are small aquatic invertebrates that have been used in aging studies for nearly 100 years and are now re-emerging as a modern model system. To provide a baseline to evaluate genetic responses to interventions that change health throughout lifespan and a framework for new hypotheses about the molecular genetic mechanisms of aging, we examined the transcriptome of an asexual female lineage of the rotifer Brachionus manjavacas at five life stages: eggs, neonates, and early-, late-, and post-reproductive adults.
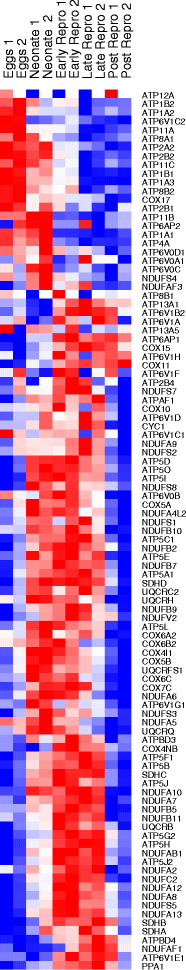
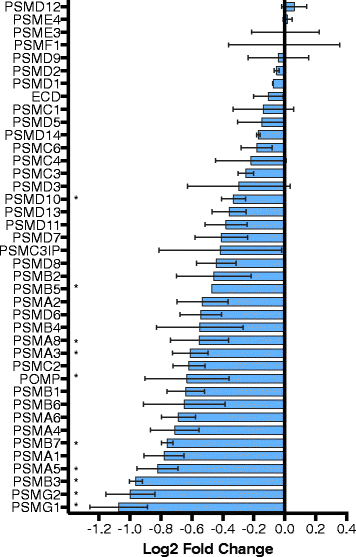
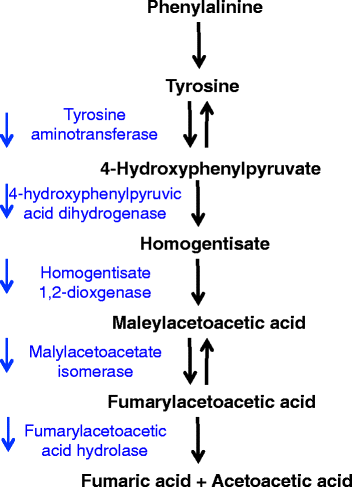
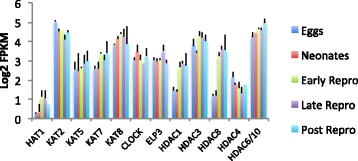
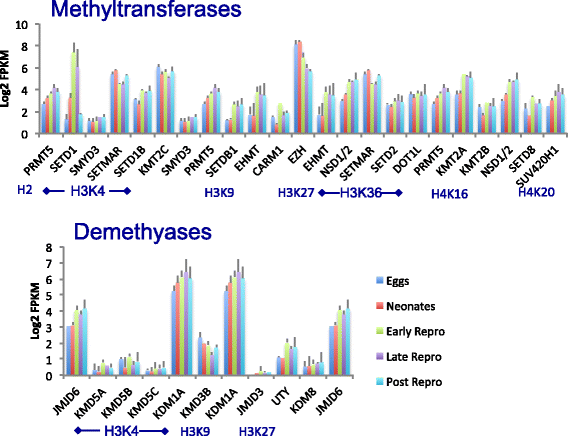
Results: There are widespread shifts in gene expression over the lifespan of B. manjavacas; the largest change occurs between neonates and early reproductive adults and is characterized by down-regulation of developmental genes and up-regulation of genes involved in reproduction. The expression profile of post-reproductive adults was distinct from that of other life stages. While few genes were significantly differentially expressed in the late- to post-reproductive transition, gene set enrichment analysis revealed multiple down-regulated pathways in metabolism, maintenance and repair, and proteostasis, united by genes involved in mitochondrial function and oxidative phosphorylation.
Conclusions: This study provides the first examination of changes in gene expression over lifespan in rotifers. We detected differential expression of many genes with human orthologs that are absent in Drosophila and C. elegans, highlighting the potential of the rotifer model in aging studies. Our findings suggest that small but coordinated changes in expression of many genes in pathways that integrate diverse functions drive the aging process. The observation of simultaneous declines in expression of genes in multiple pathways may have consequences for health and longevity not detected by single- or multi-gene knockdown in otherwise healthy animals. Investigation of subtle but genome-wide change in these pathways during aging is an important area for future study.
Keywords: Aging; Monogonont; RNA-Seq; Rotifer; Transcriptome.
Figures







References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

