ddClone: joint statistical inference of clonal populations from single cell and bulk tumour sequencing data
- PMID: 28249593
- PMCID: PMC5333399
- DOI: 10.1186/s13059-017-1169-3
ddClone: joint statistical inference of clonal populations from single cell and bulk tumour sequencing data
Abstract
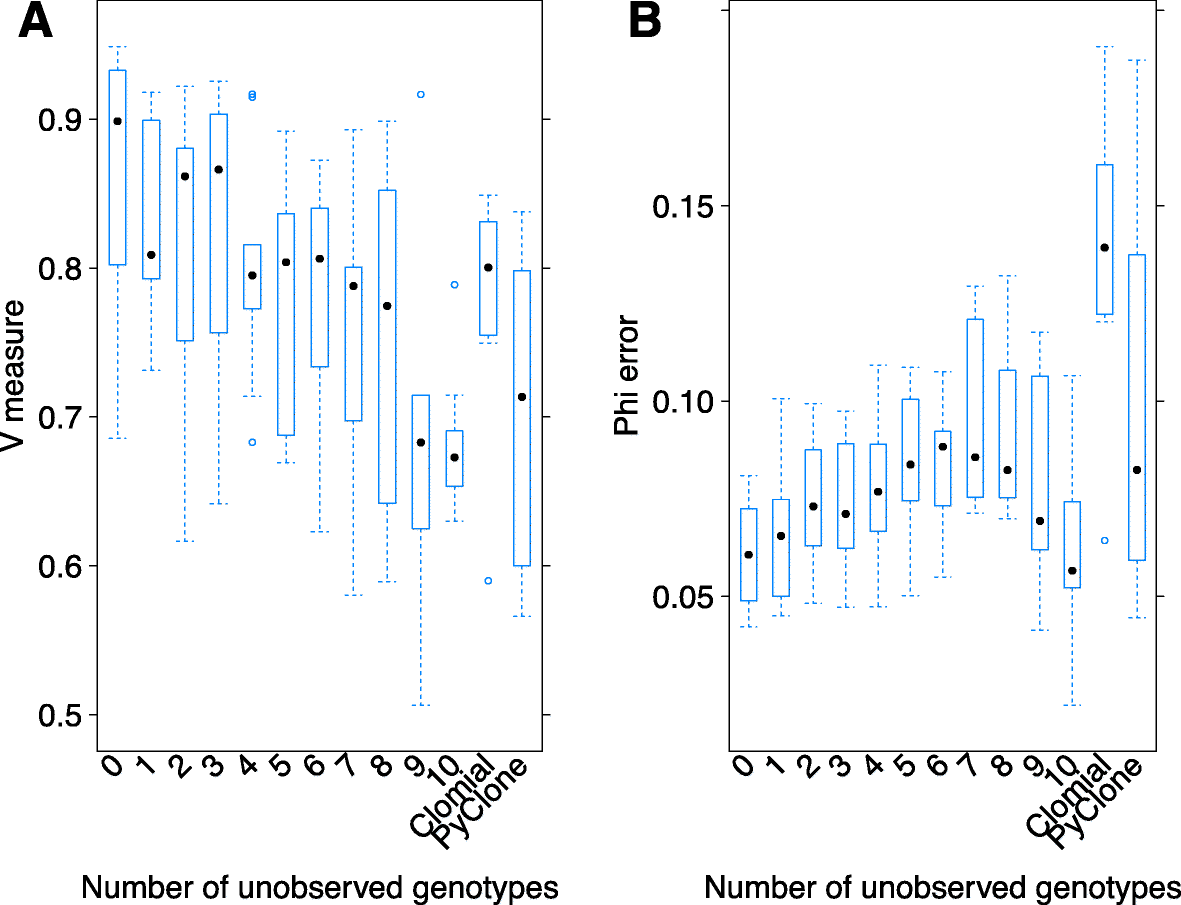
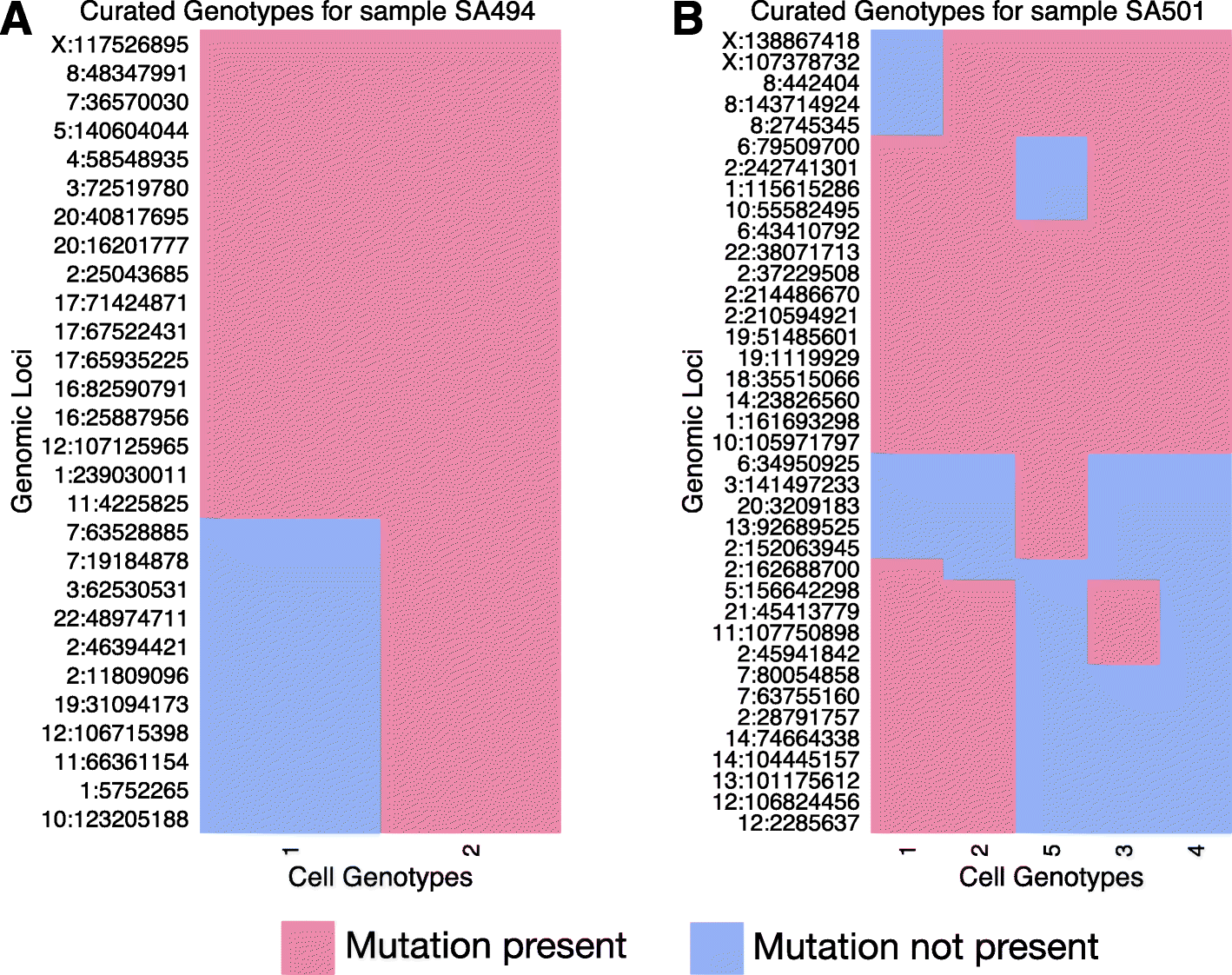
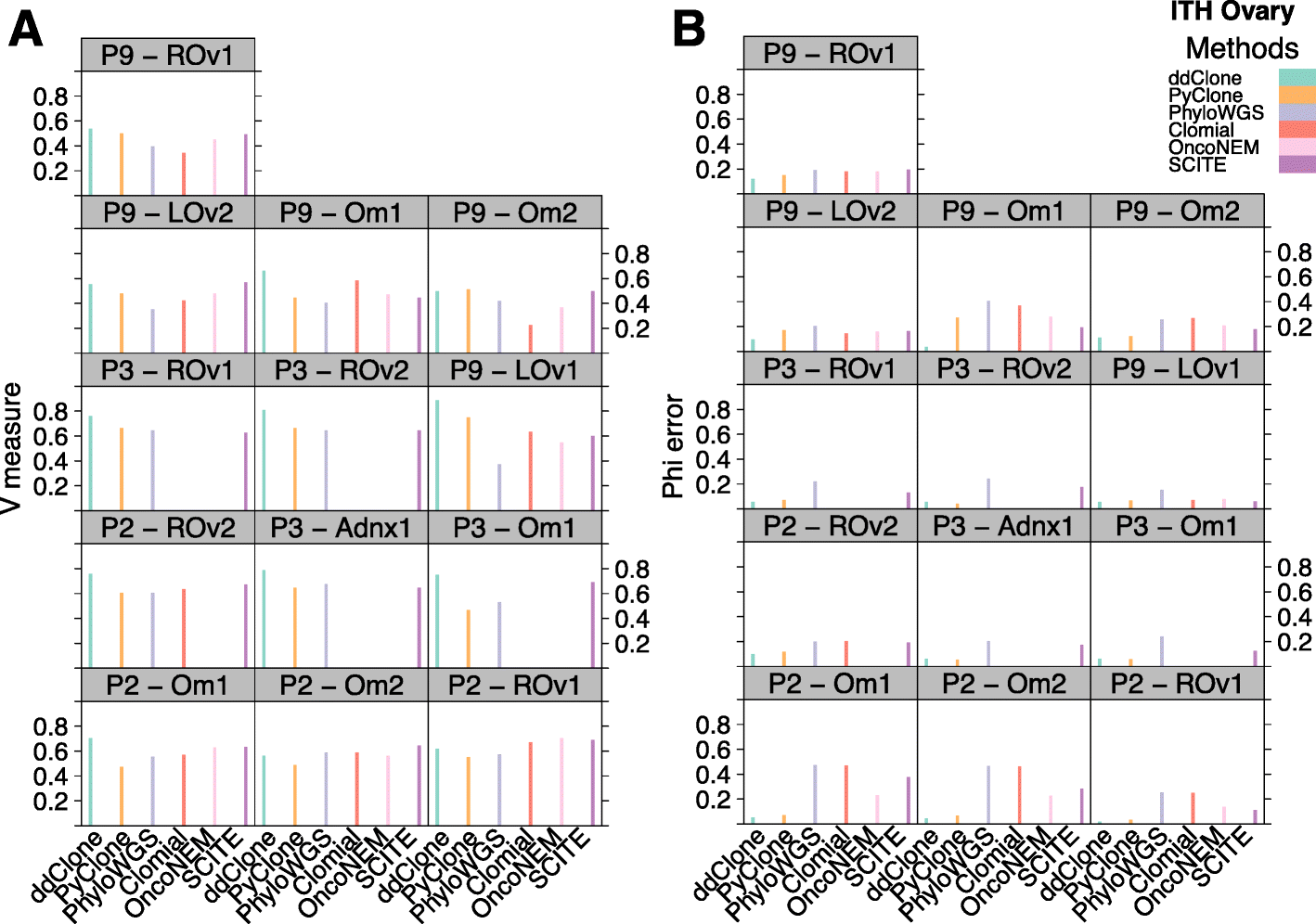
Next-generation sequencing (NGS) of bulk tumour tissue can identify constituent cell populations in cancers and measure their abundance. This requires computational deconvolution of allelic counts from somatic mutations, which may be incapable of fully resolving the underlying population structure. Single cell sequencing (SCS) is a more direct method, although its replacement of NGS is impeded by technical noise and sampling limitations. We propose ddClone, which analytically integrates NGS and SCS data, leveraging their complementary attributes through joint statistical inference. We show on real and simulated datasets that ddClone produces more accurate results than can be achieved by either method alone.
Keywords: Chinese restaurant process; Clonal evolution; Distance dependent; Intra-tumour heterogeneity; Joint probabilistic model; Next-generation sequencing; Single cell sequencing.
Figures









References
-
- Roth A, Ding J, Morin R, Crisan A, Ha G, Giuliany R, Bashashati A, Hirst M, Turashvili G, Oloumi A, et al. JointSNVMix: a probabilistic model for accurate detection of somatic mutations in normal/tumour paired next-generation sequencing data. Bioinformatics. 2012;28(7):907–13. doi: 10.1093/bioinformatics/bts053. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

