Individualized Breast Cancer Characterization through Single-Cell Analysis of Tumor and Adjacent Normal Cells
- PMID: 28249895
- PMCID: PMC5441682
- DOI: 10.1158/0008-5472.CAN-16-3308
Individualized Breast Cancer Characterization through Single-Cell Analysis of Tumor and Adjacent Normal Cells
Abstract
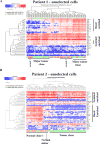
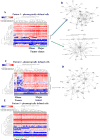
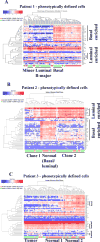
There is a need to individualize assays for tumor molecular phenotyping, given variations in the differentiation status of tumor and normal tissues in different patients. To address this, we performed single-cell genomics of breast tumors and adjacent normal cells propagated for a short duration under growth conditions that enable epithelial reprogramming. Cells analyzed were either unselected for a specific subpopulation or phenotypically defined as undifferentiated and highly clonogenic ALDH+/CD49f+/EpCAM+ luminal progenitors, which express both basal cell and luminal cell-enriched genes. We analyzed 420 tumor cells and 284 adjacent normal cells for expression of 93 genes that included a PAM50-intrinsic subtype classifier and stemness-related genes. ALDH+/CD49f+/EpCAM+ tumor and normal cells clustered differently compared with unselected tumor and normal cells. PAM50 gene-set analyses of ALDH+/CD49f+/EpCAM+ populations efficiently identified major and minor clones of tumor cells, with the major clone resembling clinical parameters of the tumor. Similarly, a stemness-associated gene set identified clones with divergent stemness pathway activation within the same tumor. This refined expression profiling technique distinguished genes truly deregulated in cancer from genes that identify cellular precursors of tumors. Collectively, the assays presented here enable more precise identification of cancer-deregulated genes, allow for early identification of therapeutically targetable tumor cell subpopulations, and ultimately provide a refinement of precision therapeutics for cancer treatment. Cancer Res; 77(10); 2759-69. ©2017 AACR.
©2017 American Association for Cancer Research.
Conflict of interest statement
Figures




References
-
- Paik S, Shak S, Tang G, Kim C, Baker J, Cronin M, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351(27):2817–26. - PubMed
-
- Prat A, Bianchini G, Thomas M, Belousov A, Cheang MC, Koehler A, et al. Research-based PAM50 subtype predictor identifies higher responses and improved survival outcomes in HER2-positive breast cancer in the NOAH study. Clin Cancer Res. 2014;20(2):511–21. - PubMed
-
- van ’t Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415(6871):530–6. - PubMed
-
- Drukker CA, Elias SG, Nijenhuis MV, Wesseling J, Bartelink H, Elkhuizen P, et al. Gene expression profiling to predict the risk of locoregional recurrence in breast cancer: a pooled analysis. Breast Cancer Res Treat. 2014;148(3):599–613. - PubMed
-
- Cardoso F, van’t Veer LJ, Bogaerts J, Slaets L, Viale G, Delaloge S, et al. 70-Gene Signature as an Aid to Treatment Decisions in Early-Stage Breast Cancer. N Engl J Med. 2016;375(8):717–29. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

