The Pivotal Role of Protein Phosphorylation in the Control of Yeast Central Metabolism
- PMID: 28250014
- PMCID: PMC5386872
- DOI: 10.1534/g3.116.037218
The Pivotal Role of Protein Phosphorylation in the Control of Yeast Central Metabolism
Abstract
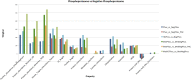
Protein phosphorylation is the most frequent eukaryotic post-translational modification and can act as either a molecular switch or rheostat for protein functions. The deliberate manipulation of protein phosphorylation has great potential for regulating specific protein functions with surgical precision, rather than the gross effects gained by the over/underexpression or complete deletion of a protein-encoding gene. In order to assess the impact of phosphorylation on central metabolism, and thus its potential for biotechnological and medical exploitation, a compendium of highly confident protein phosphorylation sites (p-sites) for the model organism Saccharomyces cerevisiae has been analyzed together with two more datasets from the fungal pathogen Candida albicans Our analysis highlights the global properties of the regulation of yeast central metabolism by protein phosphorylation, where almost half of the enzymes involved are subject to this sort of post-translational modification. These phosphorylated enzymes, compared to the nonphosphorylated ones, are more abundant, regulate more reactions, have more protein-protein interactions, and a higher fraction of them are ubiquitinated. The p-sites of metabolic enzymes are also more conserved than the background p-sites, and hundreds of them have the potential for regulating metabolite production. All this integrated information has allowed us to prioritize thousands of p-sites in terms of their potential phenotypic impact. This multi-source compendium should enable the design of future high-throughput (HTP) mutation studies to identify key molecular switches/rheostats for the manipulation of not only the metabolism of yeast, but also that of many other biotechnologically and medically important fungi and eukaryotes.
Keywords: comparative phosphoproteomics; metabolism; phosphorylation; yeast.
Copyright © 2017 Vlastaridis et al.
Figures
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
