Genotype Imputation To Improve the Cost-Efficiency of Genomic Selection in Farmed Atlantic Salmon
- PMID: 28250015
- PMCID: PMC5386885
- DOI: 10.1534/g3.117.040717
Genotype Imputation To Improve the Cost-Efficiency of Genomic Selection in Farmed Atlantic Salmon
Abstract
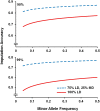
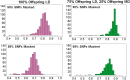
Genomic selection uses genome-wide marker information to predict breeding values for traits of economic interest, and is more accurate than pedigree-based methods. The development of high density SNP arrays for Atlantic salmon has enabled genomic selection in selective breeding programs, alongside high-resolution association mapping of the genetic basis of complex traits. However, in sibling testing schemes typical of salmon breeding programs, trait records are available on many thousands of fish with close relationships to the selection candidates. Therefore, routine high density SNP genotyping may be prohibitively expensive. One means to reducing genotyping cost is the use of genotype imputation, where selected key animals (e.g., breeding program parents) are genotyped at high density, and the majority of individuals (e.g., performance tested fish and selection candidates) are genotyped at much lower density, followed by imputation to high density. The main objectives of the current study were to assess the feasibility and accuracy of genotype imputation in the context of a salmon breeding program. The specific aims were: (i) to measure the accuracy of genotype imputation using medium (25 K) and high (78 K) density mapped SNP panels, by masking varying proportions of the genotypes and assessing the correlation between the imputed genotypes and the true genotypes; and (ii) to assess the efficacy of imputed genotype data in genomic prediction of key performance traits (sea lice resistance and body weight). Imputation accuracies of up to 0.90 were observed using the simple two-generation pedigree dataset, and moderately high accuracy (0.83) was possible even with very low density SNP data (∼250 SNPs). The performance of genomic prediction using imputed genotype data was comparable to using true genotype data, and both were superior to pedigree-based prediction. These results demonstrate that the genotype imputation approach used in this study can provide a cost-effective method for generating robust genome-wide SNP data for genomic prediction in Atlantic salmon. Genotype imputation approaches are likely to form a critical component of cost-efficient genomic selection programs to improve economically important traits in aquaculture.
Keywords: GenPred; Genomic Selection; Shared Data Resources; aquaculture; disease resistance; imputation.
Copyright © 2017 Tsai et al.
Figures

References
-
- Berry D. P., Kearney J. F., 2011. Imputation of genotypes from low- to high-density genotyping platforms and implications for genomic selection. Animal 5: 1162–1169. - PubMed
-
- Correa K., Lhorente J. P., Bassini L., Lopez M. E., Di Genova A., et al. , 2016. Genome wide association study for resistance to Caligus rogercresseyi in Atlantic salmon (Salmo salar L.) using a 50K SNP genotyping array. Aquaculture (in press).
Publication types
MeSH terms
Grants and funding
- BBS/E/D/20211550/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/M028321/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBS/E/D/20211554/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/J004235/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/J004324/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBS/E/D/20211553/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/H022007/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- MR/M000370/1/MRC_/Medical Research Council/United Kingdom
- BB/J004243/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBS/E/D/20211551/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/N024044/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
