Construction of a High-Density American Cranberry (Vaccinium macrocarpon Ait.) Composite Map Using Genotyping-by-Sequencing for Multi-pedigree Linkage Mapping
- PMID: 28250016
- PMCID: PMC5386866
- DOI: 10.1534/g3.116.037556
Construction of a High-Density American Cranberry (Vaccinium macrocarpon Ait.) Composite Map Using Genotyping-by-Sequencing for Multi-pedigree Linkage Mapping
Abstract
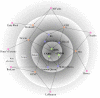
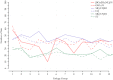
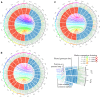
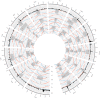
The American cranberry (Vaccinium macrocarpon Ait.) is a recently domesticated, economically important, fruit crop with limited molecular resources. New genetic resources could accelerate genetic gain in cranberry through characterization of its genomic structure and by enabling molecular-assisted breeding strategies. To increase the availability of cranberry genomic resources, genotyping-by-sequencing (GBS) was used to discover and genotype thousands of single nucleotide polymorphisms (SNPs) within three interrelated cranberry full-sib populations. Additional simple sequence repeat (SSR) loci were added to the SNP datasets and used to construct bin maps for the parents of the populations, which were then merged to create the first high-density cranberry composite map containing 6073 markers (5437 SNPs and 636 SSRs) on 12 linkage groups (LGs) spanning 1124 cM. Interestingly, higher rates of recombination were observed in maternal than paternal gametes. The large number of markers in common (mean of 57.3) and the high degree of observed collinearity (mean Pair-wise Spearman rank correlations >0.99) between the LGs of the parental maps demonstrates the utility of GBS in cranberry for identifying polymorphic SNP loci that are transferable between pedigrees and populations in future trait-association studies. Furthermore, the high-density of markers anchored within the component maps allowed identification of segregation distortion regions, placement of centromeres on each of the 12 LGs, and anchoring of genomic scaffolds. Collectively, the results represent an important contribution to the current understanding of cranberry genomic structure and to the availability of molecular tools for future genetic research and breeding efforts in cranberry.
Keywords: Vaccinium; centromere region; genetic map; simple sequence repeat; single nucleotide polymorphism.
Copyright © 2017 Schlautman et al.
Figures
Similar articles
-
Exploiting genotyping by sequencing to characterize the genomic structure of the American cranberry through high-density linkage mapping.BMC Genomics. 2016 Jun 13;17:451. doi: 10.1186/s12864-016-2802-3. BMC Genomics. 2016. PMID: 27295982 Free PMC article.
-
Construction of High Density Sweet Cherry (Prunus avium L.) Linkage Maps Using Microsatellite Markers and SNPs Detected by Genotyping-by-Sequencing (GBS).PLoS One. 2015 May 26;10(5):e0127750. doi: 10.1371/journal.pone.0127750. eCollection 2015. PLoS One. 2015. PMID: 26011256 Free PMC article.
-
Development and validation of 697 novel polymorphic genomic and EST-SSR markers in the American cranberry (Vaccinium macrocarpon Ait.).Molecules. 2015 Jan 27;20(2):2001-13. doi: 10.3390/molecules20022001. Molecules. 2015. PMID: 25633331 Free PMC article.
-
Using next-generation sequencing approaches to isolate simple sequence repeat (SSR) loci in the plant sciences.Am J Bot. 2012 Feb;99(2):193-208. doi: 10.3732/ajb.1100394. Epub 2011 Dec 20. Am J Bot. 2012. PMID: 22186186 Review.
-
Large SNP arrays for genotyping in crop plants.J Biosci. 2012 Nov;37(5):821-8. doi: 10.1007/s12038-012-9225-3. J Biosci. 2012. PMID: 23107918 Review.
Cited by
-
Massive phenotyping of multiple cranberry populations reveals novel QTLs for fruit anthocyanin content and other important chemical traits.Mol Genet Genomics. 2018 Dec;293(6):1379-1392. doi: 10.1007/s00438-018-1464-z. Epub 2018 Jul 2. Mol Genet Genomics. 2018. PMID: 29967963 Free PMC article.
-
Construction of a high density genetic linkage map to define the locus conferring seedlessness from Mukaku Kishu mandarin.Front Plant Sci. 2023 Feb 14;14:1087023. doi: 10.3389/fpls.2023.1087023. eCollection 2023. Front Plant Sci. 2023. PMID: 36875618 Free PMC article.
-
High-Resolution Linkage Map and QTL Analyses of Fruit Firmness in Autotetraploid Blueberry.Front Plant Sci. 2020 Nov 16;11:562171. doi: 10.3389/fpls.2020.562171. eCollection 2020. Front Plant Sci. 2020. PMID: 33304360 Free PMC article.
-
Pacbio Sequencing Reveals Identical Organelle Genomes between American Cranberry (Vaccinium macrocarpon Ait.) and a Wild Relative.Genes (Basel). 2019 Apr 10;10(4):291. doi: 10.3390/genes10040291. Genes (Basel). 2019. PMID: 30974783 Free PMC article.
-
Contrasting a reference cranberry genome to a crop wild relative provides insights into adaptation, domestication, and breeding.PLoS One. 2022 Mar 7;17(3):e0264966. doi: 10.1371/journal.pone.0264966. eCollection 2022. PLoS One. 2022. PMID: 35255111 Free PMC article.
References
-
- Bloom J. C., Holland J. B., 2012. Genomic localization of the maize cross-incompatibility gene, Gametophyte factor 1 (ga1). Maydica 56: 379–387.
-
- Camp W., 1945. The North American blueberries with notes on other groups of Vacciniaceae. Brittonia 5: 203–275.
-
- Cane J. H., 2009. Pollen viability and pollen tube attrition in cranberry (Vaccinium macrocarpon Aiton). Acta Hortic. 810(2): 563–566.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
