Chloroplast Preproteins Bind to the Dimer Interface of the Toc159 Receptor during Import
- PMID: 28250068
- PMCID: PMC5373065
- DOI: 10.1104/pp.16.01952
Chloroplast Preproteins Bind to the Dimer Interface of the Toc159 Receptor during Import
Abstract
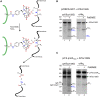
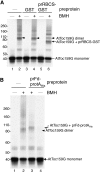
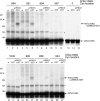
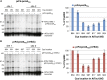
Most chloroplast proteins are synthesized in the cytosol as higher molecular weight preproteins and imported via the translocons in the outer (TOC) and inner (TIC) envelope membranes of chloroplasts. Toc159 functions as a primary receptor and directly binds preproteins through its dimeric GTPase domain. As a first step toward a molecular understanding of how Toc159 mediates preprotein import, we mapped the preprotein-binding regions on the Toc159 GTPase domain (Toc159G) of pea (Pisum sativum) using cleavage by bound preproteins conjugated with the artificial protease FeBABE and cysteine-cysteine cross-linking. Our results show that residues at the dimer interface and the switch II region of Toc159G are in close proximity to preproteins. The mature portion of preproteins was observed preferentially at the dimer interface, whereas the transit peptide was found at both regions equally. Chloroplasts from transgenic plants expressing engineered Toc159 with a cysteine placed at the dimer interface showed increased cross-linking to bound preproteins. Our data suggest that, during preprotein import, the Toc159G dimer disengages and the dimer interface contacts translocating preproteins, which is consistent with a model in which conformational changes induced by dimer-monomer conversion in Toc159 play a direct role in facilitating preprotein import.
© 2017 American Society of Plant Biologists. All Rights Reserved.
Figures



Similar articles
-
Initial binding of preproteins involving the Toc159 receptor can be bypassed during protein import into chloroplasts.Plant Physiol. 2000 Mar;122(3):813-22. doi: 10.1104/pp.122.3.813. Plant Physiol. 2000. PMID: 10712545 Free PMC article.
-
The targeting of the atToc159 preprotein receptor to the chloroplast outer membrane is mediated by its GTPase domain and is regulated by GTP.J Cell Biol. 2002 Dec 9;159(5):833-43. doi: 10.1083/jcb.200208017. Epub 2002 Dec 9. J Cell Biol. 2002. PMID: 12473690 Free PMC article.
-
A split-ubiquitin yeast two-hybrid screen to examine the substrate specificity of atToc159 and atToc132, two Arabidopsis chloroplast preprotein import receptors.PLoS One. 2014 Apr 15;9(4):e95026. doi: 10.1371/journal.pone.0095026. eCollection 2014. PLoS One. 2014. PMID: 24736607 Free PMC article.
-
Protein transport in organelles: The Toc complex way of preprotein import.FEBS J. 2009 Mar;276(5):1156-65. doi: 10.1111/j.1742-4658.2009.06873.x. FEBS J. 2009. PMID: 19187236 Review.
-
New insights into the mechanism of chloroplast protein import and its integration with protein quality control, organelle biogenesis and development.J Mol Biol. 2015 Mar 13;427(5):1038-1060. doi: 10.1016/j.jmb.2014.08.016. Epub 2014 Aug 28. J Mol Biol. 2015. PMID: 25174336 Free PMC article. Review.
Cited by
-
Origins, function, and regulation of the TOC-TIC general protein import machinery of plastids.J Exp Bot. 2020 Feb 19;71(4):1226-1238. doi: 10.1093/jxb/erz517. J Exp Bot. 2020. PMID: 31730153 Free PMC article.
-
Deciphering the Molecular Mechanisms of Chilling Tolerance in Lsi1-Overexpressing Rice.Int J Mol Sci. 2022 Apr 23;23(9):4667. doi: 10.3390/ijms23094667. Int J Mol Sci. 2022. PMID: 35563058 Free PMC article.
-
Targeted Control of Chloroplast Quality to Improve Plant Acclimation: From Protein Import to Degradation.Front Plant Sci. 2019 Jul 25;10:958. doi: 10.3389/fpls.2019.00958. eCollection 2019. Front Plant Sci. 2019. PMID: 31402924 Free PMC article. Review.
References
-
- Ahmadian MR, Stege P, Scheffzek K, Wittinghofer A (1997) Confirmation of the arginine-finger hypothesis for the GAP-stimulated GTP-hydrolysis reaction of Ras. Nat Struct Biol 4: 686–689 - PubMed
-
- Bauer J, Chen K, Hiltbunner A, Wehrli E, Eugster M, Schnell D, Kessler F (2000) The major protein import receptor of plastids is essential for chloroplast biogenesis. Nature 403: 203–207 - PubMed
-
- Brunger AT. (2007) Version 1.2 of the Crystallography and NMR system. Nat Protoc 2: 2728–2733 - PubMed
-
- Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr 54: 905–921 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

