Transcription Factor Functional Protein-Protein Interactions in Plant Defense Responses
- PMID: 28250372
- PMCID: PMC5302731
- DOI: 10.3390/proteomes2010085
Transcription Factor Functional Protein-Protein Interactions in Plant Defense Responses
Abstract
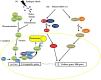
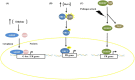
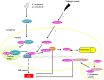
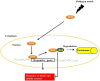
Responses to biotic stress in plants lead to dramatic reprogramming of gene expression, favoring stress responses at the expense of normal cellular functions. Transcription factors are master regulators of gene expression at the transcriptional level, and controlling the activity of these factors alters the transcriptome of the plant, leading to metabolic and phenotypic changes in response to stress. The functional analysis of interactions between transcription factors and other proteins is very important for elucidating the role of these transcriptional regulators in different signaling cascades. In this review, we present an overview of protein-protein interactions for the six major families of transcription factors involved in plant defense: basic leucine zipper containing domain proteins (bZIP), amino-acid sequence WRKYGQK (WRKY), myelocytomatosis related proteins (MYC), myeloblastosis related proteins (MYB), APETALA2/ ETHYLENE-RESPONSIVE ELEMENT BINDING FACTORS (AP2/EREBP) and no apical meristem (NAM), Arabidopsis transcription activation factor (ATAF), and cup-shaped cotyledon (CUC) (NAC). We describe the interaction partners of these transcription factors as molecular responses during pathogen attack and the key components of signal transduction pathways that take place during plant defense responses. These interactions determine the activation or repression of response pathways and are crucial to understanding the regulatory networks that modulate plant defense responses.
Keywords: biotic stress; signaling cascades; transcription factor.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
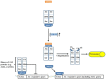
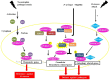

References
-
- Van Verk M.C., Gatz C., Linthorst H.J.M. Transcriptional regulation of plant defense responses. Adv. Bot. Res. 2009;51:397–438. doi: 10.1016/S0065-2296(09)51010-5. - DOI
-
- Chen W., Provart N.J., Glazebrook J., Katagiri F., Chang H.S., Eulgem T., Mauch F., Luan S., Zou G., Whitham S.A., et al. Expression profile matrix of Arabidopsis transcription factor genes suggests their putative functions in response to environmental stresses. Plant Cell. 2002;14:559–574. doi: 10.1105/tpc.010410. - DOI - PMC - PubMed
-
- Riechmann J.L. In: Transcription Factors of Arabidopsis and Rice: A Genomic Perspective in Regulation of Transcription in Plants. Grasser K.D., editor. Volume 29. Blackwell Publishing; Oxford, UK: 2006. pp. 28–53. Chapter 2.
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

