Essential role for the transcription factor Bhlhe41 in regulating the development, self-renewal and BCR repertoire of B-1a cells
- PMID: 28250425
- PMCID: PMC5363839
- DOI: 10.1038/ni.3694
Essential role for the transcription factor Bhlhe41 in regulating the development, self-renewal and BCR repertoire of B-1a cells
Abstract
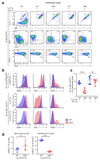
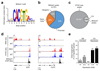
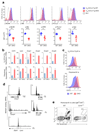
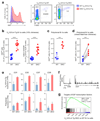
Innate-like B-1a cells provide a first line of defense against pathogens, yet little is known about their transcriptional control. Here we identified an essential role for the transcription factor Bhlhe41, with a lesser contribution by Bhlhe40, in controlling B-1a cell differentiation. Bhlhe41-/-Bhlhe40-/- B-1a cells were present at much lower abundance than were their wild-type counterparts. Mutant B-1a cells exhibited an abnormal cell-surface phenotype and altered B cell receptor (BCR) repertoire exemplified by loss of the phosphatidylcholine-specific VH12Vκ4 BCR. Expression of a pre-rearranged VH12Vκ4 BCR failed to 'rescue' the mutant phenotype and revealed enhanced proliferation accompanied by increased cell death. Bhlhe41 directly repressed the expression of cell-cycle regulators and inhibitors of BCR signaling while enabling pro-survival cytokine signaling. Thus, Bhlhe41 controls the development, BCR repertoire and self-renewal of B-1a cells.
Conflict of interest statement
The authors declare no competing financial interests.
Figures



Comment in
-
To B-1 or not to B-1.Nat Immunol. 2017 Mar 22;18(4):365-366. doi: 10.1038/ni.3715. Nat Immunol. 2017. PMID: 28323267 No abstract available.
-
Regulation of B-1a cells: another novel function of the basic helix-loop-helix transcriptional regulator BHLHE41.Cell Mol Immunol. 2017 Oct;14(10):802-804. doi: 10.1038/cmi.2017.75. Epub 2017 Sep 4. Cell Mol Immunol. 2017. PMID: 29026219 Free PMC article. No abstract available.
References
-
- Baumgarth N. The double life of a B-1 cell: self-reactivity selects for protective effector functions. Nat Rev Immunol. 2011;11:34–46. - PubMed
-
- Casola S, et al. B cell receptor signal strength determines B cell fate. Nat Immunol. 2004;5:317–327. - PubMed
-
- Hardy RR, Hayakawa K. B cell development pathways. Annu Rev Immunol. 2001;19:595–621. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

