Transcriptomic features of Pecten maximus oocyte quality and maturation
- PMID: 28253290
- PMCID: PMC5333834
- DOI: 10.1371/journal.pone.0172805
Transcriptomic features of Pecten maximus oocyte quality and maturation
Abstract
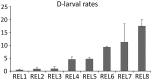
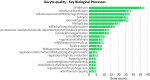
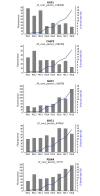
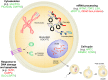
The king scallop Pecten maximus is a high valuable species of great interest in Europe for both fishery and aquaculture. Notably, there has been an increased investment to produce seed for enhancement programmes of wild scallop populations. However, hatchery production is a relatively new industry and it is still underdeveloped. Major hurdles are spawning control and gamete quality. In the present study, a total of 14 scallops were sampled in the bay of Brest (Brittany, France) to compare transcriptomic profiles of mature oocytes collected by spawning induction or by stripping. To reach such a goal, a microarray analysis was performed by using a custom 8x60K oligonucleotide microarray representing 45,488 unique scallop contigs. First we identified genes that were differentially expressed depending on oocyte quality, estimated as the potential to produce D-larvae. Secondly, we investigated the transcriptional features of both stripped and spawned oocytes. Genes coding for proteins involved in cytoskeletal dynamics, serine/threonine kinases signalling pathway, mRNA processing, response to DNA damage, apoptosis and cell-cycle appeared to be of crucial importance for both oocyte maturation and developmental competence. This study allowed us to dramatically increase the knowledge about transcriptional features of oocyte quality and maturation, as well as to propose for the first time putative molecular markers to solve a major bottleneck in scallop aquaculture.
Conflict of interest statement
Figures

References
-
- FAO (2016) Pecten maximus: species fact sheet. [Internet]. http://www.fao.org/fishery/species/3516/en (accessed 14 October 2016).
-
- Andersen S, Christophersen G, Magnesen T. Spat production of the great scallop (Pecten maximus): a roller coaster. 2011. Can J Zool;89: 579–598.
-
- Hold N, Murray LG, Kaiser MJ, Hinz H, Beaumont AR, Taylor MI. Potential effects of stock enhancement with hatchery-reared seed on genetic diversity and effective population size. Can J Fish Aquat Sci. 2012;70: 330–338.
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

