Use of a cocktail of spin traps for fingerprinting large range of free radicals in biological systems
- PMID: 28253308
- PMCID: PMC5333873
- DOI: 10.1371/journal.pone.0172998
Use of a cocktail of spin traps for fingerprinting large range of free radicals in biological systems
Abstract
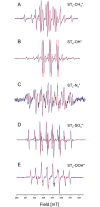
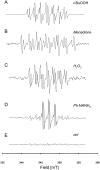
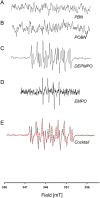
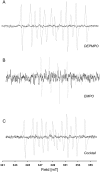
It is well established that the formation of radical species centered on various atoms is involved in the mechanism leading to the development of several diseases or to the appearance of deleterious effects of toxic molecules. The detection of free radical is possible using Electron Paramagnetic Resonance (EPR) spectroscopy and the spin trapping technique. The classical EPR spin-trapping technique can be considered as a "hypothesis-driven" approach because it requires an a priori assumption regarding the nature of the free radical in order to select the most appropriate spin-trap. We here describe a "data-driven" approach using EPR and a cocktail of spin-traps. The rationale for using this cocktail was that it would cover a wide range of biologically relevant free radicals and have a large range of hydrophilicity and lipophilicity in order to trap free radicals produced in different cellular compartments. As a proof-of-concept, we validated the ability of the system to measure a large variety of free radicals (O-, N-, C-, or S- centered) in well characterized conditions, and we illustrated the ability of the technique to unambiguously detect free radical production in cells exposed to chemicals known to be radical-mediated toxic agents.
Conflict of interest statement
Figures
Similar articles
-
Use of spin traps to detect superoxide production in living cells by electron paramagnetic resonance (EPR) spectroscopy.Methods. 2016 Oct 15;109:31-43. doi: 10.1016/j.ymeth.2016.05.001. Epub 2016 May 6. Methods. 2016. PMID: 27163864 Review.
-
Detection and characterisation of radicals using electron paramagnetic resonance (EPR) spin trapping and related methods.Methods. 2016 Oct 15;109:21-30. doi: 10.1016/j.ymeth.2016.05.013. Epub 2016 May 19. Methods. 2016. PMID: 27211009 Review.
-
Post-trapping derivatization of radical-derived EPR-silent adducts: application to free radical detection by HPLC/UV in chemical, biochemical, and biological systems and comparison with EPR spectroscopy.Anal Chem. 2012 Aug 7;84(15):6739-46. doi: 10.1021/ac301142c. Epub 2012 Jul 12. Anal Chem. 2012. PMID: 22724922
-
Kinetic analysis-based quantitation of free radical generation in EPR spin trapping.Anal Biochem. 2004 Nov 1;334(1):145-54. doi: 10.1016/j.ab.2004.07.026. Anal Biochem. 2004. PMID: 15464963
-
Development of a new EPR spin trap, DOD-8C (N-[4-dodecyloxy-2-(7'-carboxyhept-1'-yloxy)benzylidene]-N-tert-butylamine N-oxide), for the trapping of lipid radicals at a predetermined depth within biological membranes.Arch Biochem Biophys. 2005 Mar 15;435(2):336-46. doi: 10.1016/j.abb.2004.12.030. Arch Biochem Biophys. 2005. PMID: 15708377
Cited by
-
Photocatalytic Nanowires-Based Air Filter: Towards Reusable Protective Masks.Adv Funct Mater. 2020 Oct 1;30(40):2004615. doi: 10.1002/adfm.202004615. Epub 2020 Aug 7. Adv Funct Mater. 2020. PMID: 32837497 Free PMC article.
-
Membrane-specific spin trap, 5-dodecylcarbamoyl-5-N-dodecylacetamide-1-pyroline-N-oxide (diC12PO): theoretical, bioorthogonal fluorescence imaging and EPR studies.Org Biomol Chem. 2019 Sep 7;17(33):7694-7705. doi: 10.1039/c9ob01334b. Epub 2019 Jul 22. Org Biomol Chem. 2019. PMID: 31328213 Free PMC article.
-
Redox Signaling by Reactive Electrophiles and Oxidants.Chem Rev. 2018 Sep 26;118(18):8798-8888. doi: 10.1021/acs.chemrev.7b00698. Epub 2018 Aug 27. Chem Rev. 2018. PMID: 30148624 Free PMC article. Review.
-
Intracellular Metal Speciation in Streptococcus sanguinis Establishes SsaACB as Critical for Redox Maintenance.ACS Infect Dis. 2020 Jul 10;6(7):1906-1921. doi: 10.1021/acsinfecdis.0c00132. Epub 2020 May 6. ACS Infect Dis. 2020. PMID: 32329608 Free PMC article.
-
Wavelength, dose, skin type and skin model related radical formation in skin.Biophys Rev. 2021 Nov 26;13(6):1091-1100. doi: 10.1007/s12551-021-00863-0. eCollection 2021 Dec. Biophys Rev. 2021. PMID: 35047091 Free PMC article. Review.
References
-
- Halliwell B. The wanderings of a free radical. Free radical biology & medicine. 2009;46(5):531–42. Epub 2008/12/30. - PubMed
-
- Wardman P. Fluorescent and luminescent probes for measurement of oxidative and nitrosative species in cells and tissues: progress, pitfalls, and prospects. Free radical biology & medicine. 2007;43(7):995–1022. Epub 2007/09/01. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

