Specific killing of DNA damage-response deficient cells with inhibitors of poly(ADP-ribose) glycohydrolase
- PMID: 28254358
- PMCID: PMC5360195
- DOI: 10.1016/j.dnarep.2017.02.010
Specific killing of DNA damage-response deficient cells with inhibitors of poly(ADP-ribose) glycohydrolase
Abstract
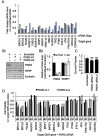
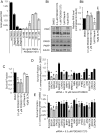
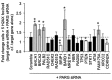
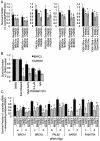
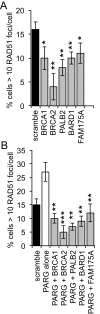
Poly(ADP-ribosylation) of proteins following DNA damage is well studied and the use of poly(ADP-ribose) polymerase (PARP) inhibitors as therapeutic agents is an exciting prospect for the treatment of many cancers. Poly(ADP-ribose) glycohydrolase (PARG) has endo- and exoglycosidase activities which can cleave glycosidic bonds, rapidly reversing the action of PARP enzymes. Like addition of poly(ADP-ribose) (PAR) by PARP, removal of PAR by PARG is also thought to be required for repair of DNA strand breaks and for continued replication at perturbed forks. Here we use siRNA to show a synthetic lethal relationship between PARG and BRCA1, BRCA2, PALB2, FAM175A (ABRAXAS) and BARD1. In addition, we demonstrate that MCF7 cells depleted of these proteins are sensitive to Gallotannin and a novel and specific PARG inhibitor PDD00017273. We confirm that PARG inhibition increases endogenous DNA damage, stalls replication forks and increases homologous recombination, and propose that it is the lack of homologous recombination (HR) proteins at PARG inhibitor-induced stalled replication forks that induces cell death. Interestingly not all genes that are synthetically lethal with PARP result in sensitivity to PARG inhibitors, suggesting that although there is overlap, the functions of PARP and PARG may not be completely identical. These data together add further evidence to the possibility that single treatment therapy with PARG inhibitors could be used for treatment of certain HR deficient tumours and provide insight into the relationship between PARP, PARG and the processes of DNA repair.
Keywords: BARD1; BRCA1; Breast cancer; FAM175A (ABRAXAS); PALB2; PARG inhibition; Synthetic lethality.
Copyright © 2017 The Authors. Published by Elsevier B.V. All rights reserved.
Figures
Similar articles
-
Inhibition of poly(ADP-ribose) glycohydrolase (PARG) specifically kills BRCA2-deficient tumor cells.Cell Cycle. 2012 Mar 1;11(5):990-7. doi: 10.4161/cc.11.5.19482. Epub 2012 Mar 1. Cell Cycle. 2012. PMID: 22333589
-
Poly(ADP-Ribose) Glycohydrolase (PARG) vs. Poly(ADP-Ribose) Polymerase (PARP) - Function in Genome Maintenance and Relevance of Inhibitors for Anti-cancer Therapy.Front Mol Biosci. 2020 Aug 28;7:191. doi: 10.3389/fmolb.2020.00191. eCollection 2020. Front Mol Biosci. 2020. PMID: 33005627 Free PMC article. Review.
-
Targeting poly(ADP-ribose) glycohydrolase to draw apoptosis codes in cancer.Biochem Pharmacol. 2019 Sep;167:163-172. doi: 10.1016/j.bcp.2019.06.004. Epub 2019 Jun 6. Biochem Pharmacol. 2019. PMID: 31176615 Review.
-
Poly(ADP-ribose) glycohydrolase mediates oxidative and excitotoxic neuronal death.Proc Natl Acad Sci U S A. 2001 Oct 9;98(21):12227-32. doi: 10.1073/pnas.211202598. Proc Natl Acad Sci U S A. 2001. PMID: 11593040 Free PMC article.
-
Selective Loss of PARG Restores PARylation and Counteracts PARP Inhibitor-Mediated Synthetic Lethality.Cancer Cell. 2018 Jun 11;33(6):1078-1093.e12. doi: 10.1016/j.ccell.2018.05.008. Cancer Cell. 2018. PMID: 29894693
Cited by
-
A novel targeted NGS panel identifies numerous homologous recombination deficiency (HRD)-associated gene mutations in addition to known BRCA mutations.Diagn Pathol. 2024 Jan 6;19(1):9. doi: 10.1186/s13000-023-01431-8. Diagn Pathol. 2024. PMID: 38184614 Free PMC article.
-
Targeting DNA repair pathway in cancer: Mechanisms and clinical application.MedComm (2020). 2021 Dec 7;2(4):654-691. doi: 10.1002/mco2.103. eCollection 2021 Dec. MedComm (2020). 2021. PMID: 34977872 Free PMC article. Review.
-
Poly (ADP) Ribose Glycohydrolase Can Be Effectively Targeted in Pancreatic Cancer.Cancer Res. 2019 Sep 1;79(17):4491-4502. doi: 10.1158/0008-5472.CAN-18-3645. Epub 2019 Jul 4. Cancer Res. 2019. PMID: 31273064 Free PMC article.
-
Targeted and Persistent 8-Oxoguanine Base Damage at Telomeres Promotes Telomere Loss and Crisis.Mol Cell. 2019 Jul 11;75(1):117-130.e6. doi: 10.1016/j.molcel.2019.04.024. Epub 2019 May 14. Mol Cell. 2019. PMID: 31101499 Free PMC article.
-
Radiosensitization with an inhibitor of poly(ADP-ribose) glycohydrolase: A comparison with the PARP1/2/3 inhibitor olaparib.DNA Repair (Amst). 2018 Jan;61:25-36. doi: 10.1016/j.dnarep.2017.11.004. Epub 2017 Nov 22. DNA Repair (Amst). 2018. PMID: 29179156 Free PMC article.
References
-
- Schreiber V., Dantzer F., Ame J.C., de Murcia G. Poly(ADP-ribose): novel functions for an old molecule. Nat. Rev. Mol. Cell Biol. 2006;7:517–528. - PubMed
-
- Benjamin R.C., Gill D.M. Poly(ADP-ribose) synthesis in vitro programmed by damaged DNA a comparison of DNA molecules containing different types of strand Breaks. J. Biol. Chem. 1980;255:10502–10508. - PubMed
-
- Ding R., Pommier Y., Kang V.H., Smulson M. Depletion of poly(ADP-ribose) polymerase by antisense RNA expression results in a delay in DNA strand break rejoining. J. Biol. Chem. 1992;267:12804–12812. - PubMed
-
- Le Page F., Schreiber V., Dherin C., De Murcia G., Boiteux S. Poly(ADP-ribose) polymerase-1(PARP-1) is required in murine cell lines for base excision repair of oxidative DNA damage in absence of DNA polymerase beta. J. Biol. Chem. 2003;278:18471–18477. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

