p38 inhibition provides anti-DNA virus immunity by regulation of USP21 phosphorylation and STING activation
- PMID: 28254948
- PMCID: PMC5379979
- DOI: 10.1084/jem.20161387
p38 inhibition provides anti-DNA virus immunity by regulation of USP21 phosphorylation and STING activation
Abstract
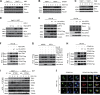
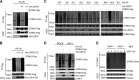
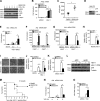
Stimulator of IFN genes (STING) is a central adaptor protein that mediates the innate immune responses to DNA virus infection. Although ubiquitination is essential for STING function, how the ubiquitination/deubiquitination system is regulated by virus infection to control STING activity remains unknown. In this study, we found that USP21 is an important deubiquitinating enzyme for STING and that it negatively regulates the DNA virus-induced production of type I interferons by hydrolyzing K27/63-linked polyubiquitin chain on STING. HSV-1 infection recruited USP21 to STING at late stage by p38-mediated phosphorylation of USP21 at Ser538. Inhibition of p38 MAPK enhanced the production of IFNs in response to virus infection and protected mice from lethal HSV-1 infection. Thus, our study reveals a critical role of p38-mediated USP21 phosphorylation in regulating STING-mediated antiviral functions and identifies p38-USP21 axis as an important pathway that DNA virus adopts to avoid innate immunity responses.
© 2017 Chen et al.
Figures







References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

