International Union of Basic and Clinical Pharmacology. CI. Structures and Small Molecule Modulators of Mammalian Adenylyl Cyclases
- PMID: 28255005
- PMCID: PMC5394921
- DOI: 10.1124/pr.116.013078
International Union of Basic and Clinical Pharmacology. CI. Structures and Small Molecule Modulators of Mammalian Adenylyl Cyclases
Abstract
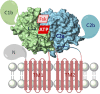
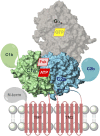
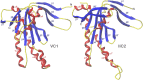
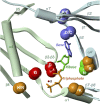
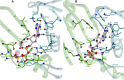
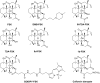
Adenylyl cyclases (ACs) generate the second messenger cAMP from ATP. Mammalian cells express nine transmembrane AC (mAC) isoforms (AC1-9) and a soluble AC (sAC, also referred to as AC10). This review will largely focus on mACs. mACs are activated by the G-protein Gαs and regulated by multiple mechanisms. mACs are differentially expressed in tissues and regulate numerous and diverse cell functions. mACs localize in distinct membrane compartments and form signaling complexes. sAC is activated by bicarbonate with physiologic roles first described in testis. Crystal structures of the catalytic core of a hybrid mAC and sAC are available. These structures provide detailed insights into the catalytic mechanism and constitute the basis for the development of isoform-selective activators and inhibitors. Although potent competitive and noncompetitive mAC inhibitors are available, it is challenging to obtain compounds with high isoform selectivity due to the conservation of the catalytic core. Accordingly, caution must be exerted with the interpretation of intact-cell studies. The development of isoform-selective activators, the plant diterpene forskolin being the starting compound, has been equally challenging. There is no known endogenous ligand for the forskolin binding site. Recently, development of selective sAC inhibitors was reported. An emerging field is the association of AC gene polymorphisms with human diseases. For example, mutations in the AC5 gene (ADCY5) cause hyperkinetic extrapyramidal motor disorders. Overall, in contrast to the guanylyl cyclase field, our understanding of the (patho)physiology of AC isoforms and the development of clinically useful drugs targeting ACs is still in its infancy.
Copyright © 2017 by The American Society for Pharmacology and Experimental Therapeutics.
Figures
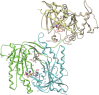

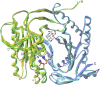
Similar articles
-
Structure, mechanism, and regulation of soluble adenylyl cyclases - similarities and differences to transmembrane adenylyl cyclases.Biochim Biophys Acta. 2014 Dec;1842(12 Pt B):2535-47. doi: 10.1016/j.bbadis.2014.08.012. Epub 2014 Sep 2. Biochim Biophys Acta. 2014. PMID: 25193033 Review.
-
Inhibitors of membranous adenylyl cyclases.Trends Pharmacol Sci. 2012 Feb;33(2):64-78. doi: 10.1016/j.tips.2011.10.006. Epub 2011 Nov 17. Trends Pharmacol Sci. 2012. PMID: 22100304 Free PMC article. Review.
-
Mammalian Nucleotidyl Cyclases and Their Nucleotide Binding Sites.Handb Exp Pharmacol. 2017;238:49-66. doi: 10.1007/164_2015_34. Handb Exp Pharmacol. 2017. PMID: 27900607 Review.
-
Structural analysis of human soluble adenylyl cyclase and crystal structures of its nucleotide complexes-implications for cyclase catalysis and evolution.FEBS J. 2014 Sep;281(18):4151-64. doi: 10.1111/febs.12913. Epub 2014 Jul 28. FEBS J. 2014. PMID: 25040695
-
Isoform selectivity of adenylyl cyclase inhibitors: characterization of known and novel compounds.J Pharmacol Exp Ther. 2013 Nov;347(2):265-75. doi: 10.1124/jpet.113.208157. Epub 2013 Sep 4. J Pharmacol Exp Ther. 2013. PMID: 24006339 Free PMC article.
Cited by
-
Protein-protein interaction-based high throughput screening for adenylyl cyclase 1 inhibitors: Design, implementation, and discovery of a novel chemotype.Front Pharmacol. 2022 Sep 6;13:977742. doi: 10.3389/fphar.2022.977742. eCollection 2022. Front Pharmacol. 2022. PMID: 36147328 Free PMC article.
-
The evolutionary conservation of eukaryotic membrane-bound adenylyl cyclase isoforms.Front Pharmacol. 2022 Sep 27;13:1009797. doi: 10.3389/fphar.2022.1009797. eCollection 2022. Front Pharmacol. 2022. PMID: 36238545 Free PMC article.
-
Direct stimulation of adenylyl cyclase 9 by the fungicide imidazole miconazole.Naunyn Schmiedebergs Arch Pharmacol. 2019 Apr;392(4):497-504. doi: 10.1007/s00210-018-01610-1. Epub 2019 Jan 3. Naunyn Schmiedebergs Arch Pharmacol. 2019. PMID: 30607468
-
Therapeutic Implications for PDE2 and cGMP/cAMP Mediated Crosstalk in Cardiovascular Diseases.Int J Mol Sci. 2020 Oct 10;21(20):7462. doi: 10.3390/ijms21207462. Int J Mol Sci. 2020. PMID: 33050419 Free PMC article. Review.
-
Unusual phototransduction via cross-motif signaling from Gq to adenylyl cyclase in intrinsically photosensitive retinalganglion cells.Proc Natl Acad Sci U S A. 2023 Jan 3;120(1):e2216599120. doi: 10.1073/pnas.2216599120. Epub 2022 Dec 29. Proc Natl Acad Sci U S A. 2023. PMID: 36584299 Free PMC article.
References
-
- Abdel-Majid RM, Leong WL, Schalkwyk LC, Smallman DS, Wong ST, Storm DR, Fine A, Dobson MJ, Guernsey DL, Neumann PE. (1998) Loss of adenylyl cyclase I activity disrupts patterning of mouse somatosensory cortex. Nat Genet 19:289–291. - PubMed
-
- Alasbahi RH, Melzig MF. (2012) Forskolin and derivatives as tools for studying the role of cAMP. Pharmazie 67:5–13. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

