Nucleophosmin Interacts with PIN2/TERF1-interacting Telomerase Inhibitor 1 (PinX1) and Attenuates the PinX1 Inhibition on Telomerase Activity
- PMID: 28255170
- PMCID: PMC5334639
- DOI: 10.1038/srep43650
Nucleophosmin Interacts with PIN2/TERF1-interacting Telomerase Inhibitor 1 (PinX1) and Attenuates the PinX1 Inhibition on Telomerase Activity
Abstract
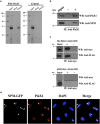
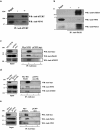
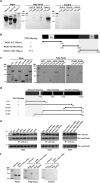
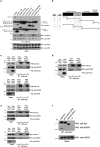
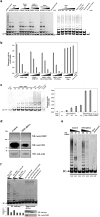
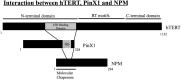
Telomerase activation and telomere maintenance are critical for cellular immortalization and transformation. PIN2/TERF1-interacting telomerase inhibitor 1 (PinX1) is a telomerase regulator and the aberrant expression of PinX1 causes telomere shortening. Identifying PinX1-interacting proteins is important for understanding telomere maintenance. We found that PinX1 directly interacts with nucleophosmin (NPM), a protein that has been shown to positively correlate with telomerase activity. We further showed that PinX1 acts as a linker in the association between NPM and hTERT, the catalytic subunit of telomerase. Additionally, the recruitment of NPM by PinX1 to the telomerase complex could partially attenuate the PinX1-mediated inhibition on telomerase activity. Taken together, our data reveal a novel mechanism that regulates telomerase activation through the interaction between NPM, PinX1 and the telomerase complex.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
Similar articles
-
The PinX1/NPM interaction associates with hTERT in early-S phase and facilitates telomerase activation.Cell Biosci. 2019 Jun 13;9:47. doi: 10.1186/s13578-019-0306-y. eCollection 2019. Cell Biosci. 2019. PMID: 31210926 Free PMC article.
-
C-terminal amino acids 290-328 of LPTS/PinX1 confer telomerase inhibition.Biochem Biophys Res Commun. 2010 Aug 6;398(4):683-9. doi: 10.1016/j.bbrc.2010.06.136. Epub 2010 Jul 8. Biochem Biophys Res Commun. 2010. PMID: 20620128
-
Characterization of interactions between PinX1 and human telomerase subunits hTERT and hTR.J Biol Chem. 2004 Dec 10;279(50):51745-8. doi: 10.1074/jbc.M408131200. Epub 2004 Sep 20. J Biol Chem. 2004. PMID: 15381700
-
PinX1: structure, regulation and its functions in cancer.Oncotarget. 2016 Oct 4;7(40):66267-66275. doi: 10.18632/oncotarget.11411. Oncotarget. 2016. PMID: 27556185 Free PMC article. Review.
-
PinX1 plays multifaceted roles in human cancers: a review and perspectives.Mol Biol Rep. 2024 Nov 17;51(1):1163. doi: 10.1007/s11033-024-10082-x. Mol Biol Rep. 2024. PMID: 39550726 Free PMC article. Review.
Cited by
-
Post-Transcriptional and Post-Translational Modifications in Telomerase Biogenesis and Recruitment to Telomeres.Int J Mol Sci. 2023 Mar 6;24(5):5027. doi: 10.3390/ijms24055027. Int J Mol Sci. 2023. PMID: 36902458 Free PMC article. Review.
-
Electrochemical determination of the activity and inhibition of telomerase based on the interaction of DNA with molybdate.Mikrochim Acta. 2019 Jan 10;186(2):96. doi: 10.1007/s00604-018-3223-6. Mikrochim Acta. 2019. PMID: 30631950
-
The PinX1/NPM interaction associates with hTERT in early-S phase and facilitates telomerase activation.Cell Biosci. 2019 Jun 13;9:47. doi: 10.1186/s13578-019-0306-y. eCollection 2019. Cell Biosci. 2019. PMID: 31210926 Free PMC article.
-
Composition and Function of Telomerase-A Polymerase Associated with the Origin of Eukaryotes.Biomolecules. 2020 Oct 8;10(10):1425. doi: 10.3390/biom10101425. Biomolecules. 2020. PMID: 33050064 Free PMC article. Review.
-
Telomerase Biogenesis and Activities from the Perspective of Its Direct Interacting Partners.Cancers (Basel). 2020 Jun 24;12(6):1679. doi: 10.3390/cancers12061679. Cancers (Basel). 2020. PMID: 32599885 Free PMC article. Review.
References
-
- Chakhparonian M. & Wellinger R. J. Telomere maintenance and DNA replication: how closely are these two connected? Trends Genet. 19, 439–446 (2003). - PubMed
-
- Colgin L. & Reddel R. Telomere biology: a new player in the end zone. Curr. Biol. 14, R901–902 (2004). - PubMed
-
- Smith S., Giriat I., Schmitt A. & de Lange T. Tankyrase, a poly(ADP-ribose) polymerase at human telomeres. Science 282, 1484–1487 (1998). - PubMed
-
- van Steensel B. & de Lange T. Control of telomere length by the human telomeric protein TRF1. Nature 385, 740–743 (1997). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

