Nutrient Sensing at the Plasma Membrane of Fungal Cells
- PMID: 28256189
- PMCID: PMC11687466
- DOI: 10.1128/microbiolspec.FUNK-0031-2016
Nutrient Sensing at the Plasma Membrane of Fungal Cells
Abstract
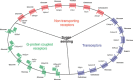
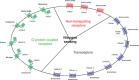
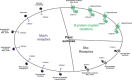
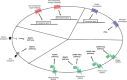
To respond to the changing environment, cells must be able to sense external conditions. This is important for many processes including growth, mating, the expression of virulence factors, and several other regulatory effects. Nutrient sensing at the plasma membrane is mediated by different classes of membrane proteins that activate downstream signaling pathways: nontransporting receptors, transceptors, classical and nonclassical G-protein-coupled receptors, and the newly defined extracellular mucin receptors. Nontransporting receptors have the same structure as transport proteins, but have lost the capacity to transport while gaining a receptor function. Transceptors are transporters that also function as a receptor, because they can rapidly activate downstream signaling pathways. In this review, we focus on these four types of fungal membrane proteins. We mainly discuss the sensing mechanisms relating to sugars, ammonium, and amino acids. Mechanisms for other nutrients, such as phosphate and sulfate, are discussed briefly. Because the model yeast Saccharomyces cerevisiae has been the most studied, especially regarding these nutrient-sensing systems, each subsection will commence with what is known in this species.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

