Limitations and challenges of genetic barcode quantification
- PMID: 28256524
- PMCID: PMC5335698
- DOI: 10.1038/srep43249
Limitations and challenges of genetic barcode quantification
Abstract
Genetic barcodes are increasingly used to track individual cells and to quantitatively assess their clonal contributions over time. Although barcode quantification relies entirely on counting sequencing reads, detailed studies about the method's accuracy are still limited. We report on a systematic investigation of the relation between barcode abundance and resulting read counts after amplification and sequencing using cell-mixtures that contain barcodes with known frequencies ("miniBulks"). We evaluated the influence of protocol modifications to identify potential sources of error and elucidate possible limitations of the quantification approach. Based on these findings we designed an advanced barcode construct (BC32) to improved barcode calling and quantification, and to ensure a sensitive detection of even highly diluted barcodes. Our results emphasize the importance of using curated barcode libraries to obtain interpretable quantitative data and underline the need for rigorous analyses of any utilized barcode library in terms of reliability and reproducibility.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

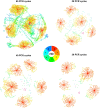



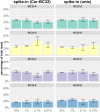

References
-
- Drize N. J. J. R. K. & Chertkov J. L. Local clonal analysis of the hematopoietic system shows that multiple small short-living clones maintain life- long hematopoiesis in reconstituted mice. Annu. Rev. Med. 47, 11–20 (1996). - PubMed
-
- Jordan C. T. & Lemischka I. R. Clonal and systemic analysis of long-term hematopoiesis in the mouse. Genes Dev. 4, 220–232 (1990). - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

