Genetic variation architecture of mitochondrial genome reveals the differentiation in Korean landrace and weedy rice
- PMID: 28256554
- PMCID: PMC5335689
- DOI: 10.1038/srep43327
Genetic variation architecture of mitochondrial genome reveals the differentiation in Korean landrace and weedy rice
Abstract
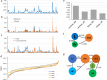
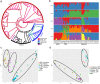
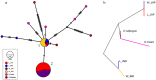
Mitochondrial genome variations have been detected despite the overall conservation of this gene content, which has been valuable for plant population genetics and evolutionary studies. Here, we describe mitochondrial variation architecture and our performance of a phylogenetic dissection of Korean landrace and weedy rice. A total of 4,717 variations across the mitochondrial genome were identified adjunct with 10 wild rice. Genetic diversity assessment revealed that wild rice has higher nucleotide diversity than landrace and/or weedy, and landrace rice has higher diversity than weedy rice. Genetic distance was suggestive of a high level of breeding between landrace and weedy rice, and the landrace showing a closer association with wild rice than weedy rice. Population structure and principal component analyses showed no obvious difference in the genetic backgrounds of landrace and weedy rice in mitochondrial genome level. Phylogenetic, population split, and haplotype network evaluations were suggestive of independent origins of the indica and japonica varieties. The origin of weedy rice is supposed to be more likely from cultivated rice rather than from wild rice in mitochondrial genome level.
Conflict of interest statement
The authors declare no competing financial interests.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

