Land-based salmon aquacultures change the quality and bacterial degradation of riverine dissolved organic matter
- PMID: 28256613
- PMCID: PMC5335611
- DOI: 10.1038/srep43739
Land-based salmon aquacultures change the quality and bacterial degradation of riverine dissolved organic matter
Abstract
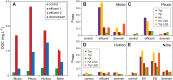
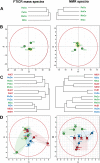
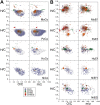
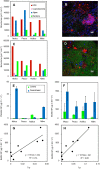
Aquacultures are of great economic importance worldwide but pollute pristine headwater streams, lakes, and estuaries. However, there are no in-depth studies of the consequences of aquacultures on dissolved organic matter (DOM) composition and structure. We performed a detailed molecular level characterization of aquaculture DOM quality and its bacterial degradation using four salmon aquacultures in Chile. Fluorescence measurements, ultrahigh-resolution mass spectrometry, and nuclear magnetic resonance spectroscopy of the DOM revealed specific and extensive molecular alterations caused by aquacultures. Aquacultures released large quantities of readily bioavailable metabolites (primarily carbohydrates and peptides/proteins, and lipids), causing the organic matter downstream of all the investigated aquacultures to deviate strongly from the highly processed, polydisperse and molecularly heterogeneous DOM found in pristine rivers. However, the upstream individual catchment DOM signatures remained distinguishable at the downstream sites. The benthic algal biovolume decreased and the bacterial biovolume and production increased downstream of the aquacultures, shifting stream ecosystems to a more heterotrophic state and thus impairing the ecosystem health. The bacterial DOM degradation rates explain the attenuation of aquaculture DOM within the subsequent stream reaches. This knowledge may aid the development of improved waste processing facilities and may help to define emission thresholds to protect sensitive stream ecosystems.
Conflict of interest statement
The authors declare no competing financial interests.
Figures


References
-
- Raymond P. A. et al.. Global carbon dioxide emissions from inland waters. Nature 503, 355–359 (2013). - PubMed
-
- Battin T. J., Besemer K., Bengtsson M. M., Romani A. M. & Packman A. I. The ecology and biogeochemistry of stream biofilms. Nat. Rev. Microbiol. 14, 251–263 (2016). - PubMed
-
- Romani A. M. et al.. Biofilm structure and function and possible implications for riverine DOC dynamics. Microb. Ecol. 47, 316–328 (2004). - PubMed
-
- Battin T. J. et al.. Biophysical controls on organic carbon fluxes in fluvial networks. Nat. Geosci. 1, 95–100 (2008).
-
- Cole J. J. et al.. Plumbing the global carbon cycle: integrating inland waters into the terrestrial carbon budget. Ecosystems 10, 171–184 (2007).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

