Three Unique Interstitial Macrophages in the Murine Lung at Steady State
- PMID: 28257233
- PMCID: PMC5516280
- DOI: 10.1165/rcmb.2016-0361OC
Three Unique Interstitial Macrophages in the Murine Lung at Steady State
Abstract
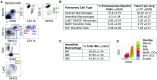
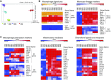
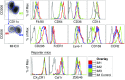
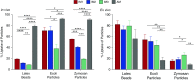
The current paradigm in macrophage biology is that some tissues mainly contain macrophages from embryonic origin, such as microglia in the brain, whereas other tissues contain postnatal-derived macrophages, such as the gut. However, in the lung and in other organs, such as the skin, there are both embryonic and postnatal-derived macrophages. In this study, we demonstrate in the steady-state lung that the mononuclear phagocyte system is comprised of three newly identified interstitial macrophages (IMs), alveolar macrophages, dendritic cells, and few extravascular monocytes. We focused on similarities and differences between the three IM subtypes, specifically, their phenotype, location, transcriptional signature, phagocytic capacity, turnover, and lack of survival dependency on fractalkine receptor, CX3CR1. Pulmonary IMs were located in the bronchial interstitium but not the alveolar interstitium. At the transcriptional level, all three IMs displayed a macrophage signature and phenotype. All IMs expressed MER proto-oncogene, tyrosine kinase, CD64, CD11b, and CX3CR1, and were further distinguished by differences in cell surface protein expression of CD206, Lyve-1, CD11c, CCR2, and MHC class II, along with the absence of Ly6C, Ly6G, and Siglec F. Most intriguingly, in addition to the lung, similar phenotypic populations of IMs were observed in other nonlymphoid organs, perhaps highlighting conserved functions throughout the body. These findings promote future research to track four distinct pulmonary macrophages and decipher the division of labor that exists between them.
Keywords: dendritic cells; interstitial macrophages; monocytes; mononuclear phagocytes; pulmonary.
Figures


References
-
- Schneider C, Nobs SP, Kurrer M, Rehrauer H, Thiele C, Kopf M. Induction of the nuclear receptor PPAR-γ by the cytokine GM-CSF is critical for the differentiation of fetal monocytes into alveolar macrophages. Nat Immunol. 2014;15:1026–1037. - PubMed
-
- Sheng J, Ruedl C, Karjalainen K. Most tissue-resident macrophages except microglia are derived from fetal hematopoietic stem cells. Immunity. 2015;43:382–393. - PubMed
-
- Scott CL, Henri S, Guilliams M. Mononuclear phagocytes of the intestine, the skin, and the lung. Immunol Rev. 2014;262:9–24. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

