Meta-evaluation of meta-analysis: ten appraisal questions for biologists
- PMID: 28257642
- PMCID: PMC5336618
- DOI: 10.1186/s12915-017-0357-7
Meta-evaluation of meta-analysis: ten appraisal questions for biologists
Abstract
Meta-analysis is a statistical procedure for analyzing the combined data from different studies, and can be a major source of concise up-to-date information. The overall conclusions of a meta-analysis, however, depend heavily on the quality of the meta-analytic process, and an appropriate evaluation of the quality of meta-analysis (meta-evaluation) can be challenging. We outline ten questions biologists can ask to critically appraise a meta-analysis. These questions could also act as simple and accessible guidelines for the authors of meta-analyses. We focus on meta-analyses using non-human species, which we term 'biological' meta-analysis. Our ten questions are aimed at enabling a biologist to evaluate whether a biological meta-analysis embodies 'mega-enlightenment', a 'mega-mistake', or something in between.
Keywords: Biological importance; Effect size; Meta-regression; Meta-research; Non-independence; Publication bias; Quantitative synthesis; Reporting bias; Statistical significance; Systematic review.
Figures



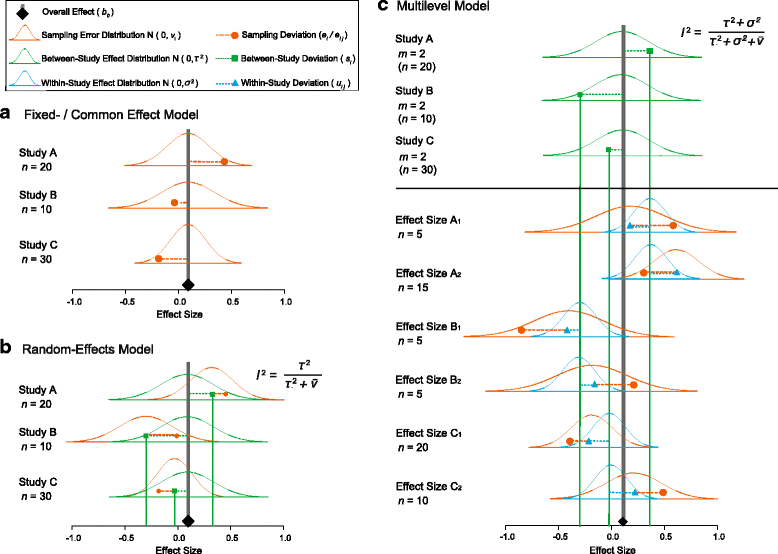
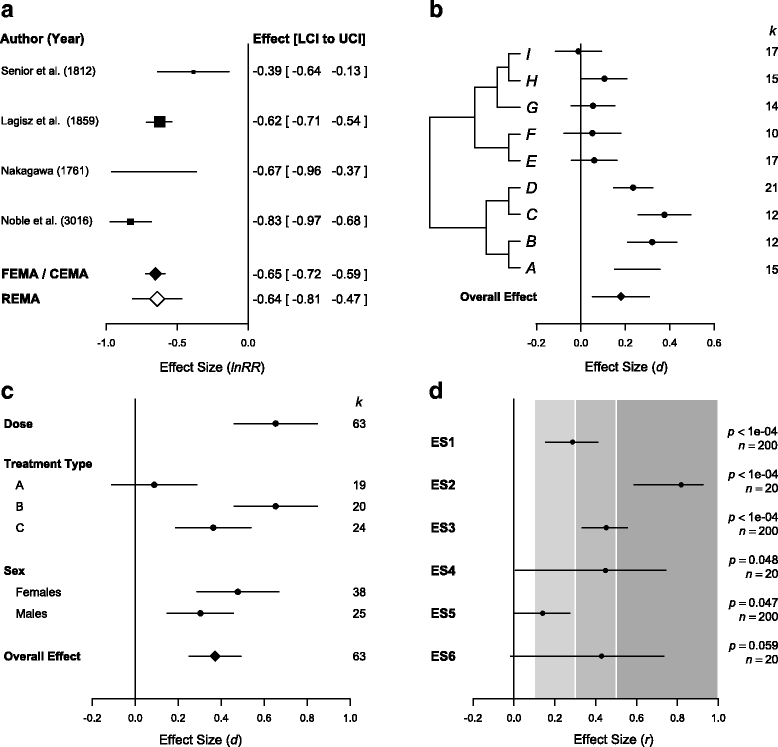
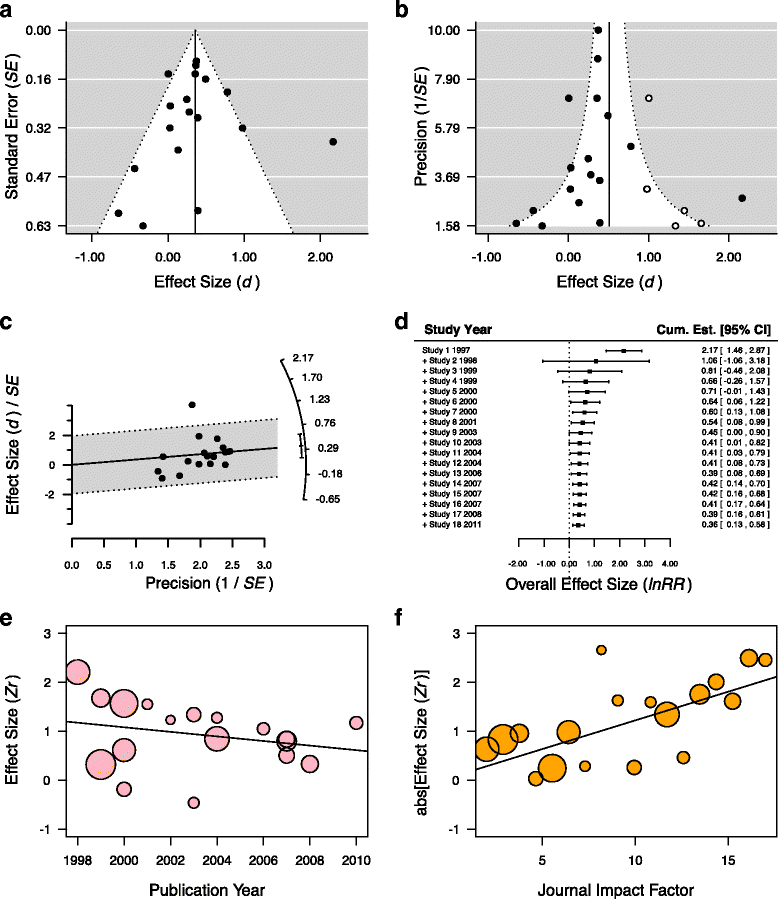
References
-
- Glass GV. Primary, secondary, and meta-analysis research. Educ Res. 1976;5:3–8. doi: 10.3102/0013189X005010003. - DOI
-
- Cooper H, Hedges LV, Valentine JC. The handbook of research synthesis and meta-analysis. New York: Russell Sage Foundation; 2009.
-
- Hedges L, Olkin I. Statistical methods for meta-analysis. New York: Academic Press; 1985.
-
- Egger M, Smith GD, Altman DG. Systematic reviews in health care: meta-analysis in context. 2. London: BMJ; 2001.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

