RNP transport in cell biology: the long and winding road
- PMID: 28258033
- PMCID: PMC5482755
- DOI: 10.1016/j.ceb.2017.02.008
RNP transport in cell biology: the long and winding road
Abstract
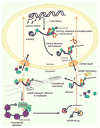
Regulation of gene expression is key determinant to cell structure and function. RNA localization, where specific mRNAs are transported to subcellular regions and then translated, is highly conserved in eukaryotes ranging from yeast to extremely specialized and polarized cells such as neurons. Messenger RNA and associated proteins (mRNP) move from the site of transcription in the nucleus to their final destination in the cytoplasm both passively through diffusion and actively via directed transport. Dysfunction of RNA localization, transport and translation machinery can lead to pathology. Single-molecule live-cell imaging techniques have revealed unique features of this journey with unprecedented resolution. In this review, we highlight key recent findings that have been made using these approaches and possible implications for spatial control of gene function.
Copyright © 2017 Elsevier Ltd. All rights reserved.
Figures

References
-
- Lecuyer E, Yoshida H, Parthasarathy N, Alm C, Babak T, Cerovina T, Hughes TR, Tomancak P, Krause HM. Global analysis of mRNA localization reveals a prominent role in organizing cellular architecture and function. Cell. 2007;131:174–187. - PubMed
-
- Lawrence JB, Singer RH. Intracellular localization of messenger RNAs for cytoskeletal proteins. Cell. 1986;45:407–415. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

