Quantifying the Importance of the Rare Biosphere for Microbial Community Response to Organic Pollutants in a Freshwater Ecosystem
- PMID: 28258138
- PMCID: PMC5377499
- DOI: 10.1128/AEM.03321-16
Quantifying the Importance of the Rare Biosphere for Microbial Community Response to Organic Pollutants in a Freshwater Ecosystem
Abstract
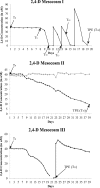
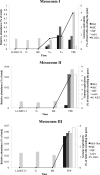
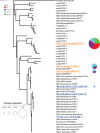
A single liter of water contains hundreds, if not thousands, of bacterial and archaeal species, each of which typically makes up a very small fraction of the total microbial community (<0.1%), the so-called "rare biosphere." How often, and via what mechanisms, e.g., clonal amplification versus horizontal gene transfer, the rare taxa and genes contribute to microbial community response to environmental perturbations represent important unanswered questions toward better understanding the value and modeling of microbial diversity. We tested whether rare species frequently responded to changing environmental conditions by establishing 20-liter planktonic mesocosms with water from Lake Lanier (Georgia, USA) and perturbing them with organic compounds that are rarely detected in the lake, including 2,4-dichlorophenoxyacetic acid (2,4-D), 4-nitrophenol (4-NP), and caffeine. The populations of the degraders of these compounds were initially below the detection limit of quantitative PCR (qPCR) or metagenomic sequencing methods, but they increased substantially in abundance after perturbation. Sequencing of several degraders (isolates) and time-series metagenomic data sets revealed distinct cooccurring alleles of degradation genes, frequently carried on transmissible plasmids, especially for the 2,4-D mesocosms, and distinct species dominating the post-enrichment microbial communities from each replicated mesocosm. This diversity of species and genes also underlies distinct degradation profiles among replicated mesocosms. Collectively, these results supported the hypothesis that the rare biosphere can serve as a genetic reservoir, which can be frequently missed by metagenomics but enables community response to changing environmental conditions caused by organic pollutants, and they provided insights into the size of the pool of rare genes and species.IMPORTANCE A single liter of water or gram of soil contains hundreds of low-abundance bacterial and archaeal species, the so called rare biosphere. The value of this astonishing biodiversity for ecosystem functioning remains poorly understood, primarily due to the fact that microbial community analysis frequently focuses on abundant organisms. Using a combination of culture-dependent and culture-independent (metagenomics) techniques, we showed that rare taxa and genes commonly contribute to the microbial community response to organic pollutants. Our findings should have implications for future studies that aim to study the role of rare species in environmental processes, including environmental bioremediation efforts of oil spills or other contaminants.
Keywords: biodegradation; community response; metagenomics; organic contaminants; rare biosphere.
Copyright © 2017 American Society for Microbiology.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

