Pseudomonas aeruginosa Aggregate Formation in an Alginate Bead Model System Exhibits In Vivo-Like Characteristics
- PMID: 28258141
- PMCID: PMC5394317
- DOI: 10.1128/AEM.00113-17
Pseudomonas aeruginosa Aggregate Formation in an Alginate Bead Model System Exhibits In Vivo-Like Characteristics
Abstract
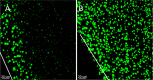
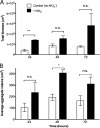
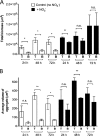
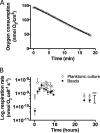
Alginate beads represent a simple and highly reproducible in vitro model system for diffusion-limited bacterial growth. In this study, alginate beads were inoculated with Pseudomonas aeruginosa and followed for up to 72 h. Confocal microscopy revealed that P. aeruginosa formed dense clusters similar in size to in vivo aggregates observed ex vivo in cystic fibrosis lungs and chronic wounds. Bacterial aggregates primarily grew in the bead periphery and decreased in size and abundance toward the center of the bead. Microsensor measurements showed that the O2 concentration decreased rapidly and reached anoxia ∼100 μm below the alginate bead surface. This gradient was relieved in beads supplemented with NO3- as an alternative electron acceptor allowing for deeper growth into the beads. A comparison of gene expression profiles between planktonic and alginate-encapsulated P. aeruginosa confirmed that the bacteria experienced hypoxic and anoxic growth conditions. Furthermore, alginate-encapsulated P. aeruginosa exhibited a lower respiration rate than the planktonic counterpart and showed a high tolerance toward antibiotics. The inoculation and growth of P. aeruginosa in alginate beads represent a simple and flexible in vivo-like biofilm model system, wherein bacterial growth exhibits central features of in vivo biofilms. This was observed by the formation of small cell aggregates in a secondary matrix with O2-limited growth, which was alleviated by the addition of NO3- as an alternative electron acceptor, and by reduced respiration rates, as well as an enhanced tolerance to antibiotic treatment.IMPORTANCEPseudomonas aeruginosa has been studied intensively for decades due to its involvement in chronic infections, such as cystic fibrosis and chronic wounds, where it forms biofilms. Much research has been dedicated to biofilm formation on surfaces; however, in chronic infections, most biofilms form small aggregates of cells not attached to a surface, but embedded in host material. In this study, bacteria were encapsulated in small alginate beads and formed aggregates similar to what is observed in chronic bacterial infections. Our findings show that aggregates are exposed to steep oxygen gradients, with zones of oxygen depletion, and that nitrate may serve as an alternative to oxygen, enabling growth in oxygen-depleted zones. This is important, as slow growth under low-oxygen conditions may render the bacteria tolerant toward antibiotics. This model provides an alternative to surface biofilm models and adds to the comprehension that biofilms do not depend on a surface for formation.
Keywords: Pseudomonas aeruginosa; antibiotics; biofilm; chronic infection; growth; model system; respiration; spatial structure.
Copyright © 2017 Sønderholm et al.
Figures




References
-
- Hoiby N. 1977. Pseudomonas aeruginosa infection in cystic fibrosis. Diagnostic and prognostic significance of pseudomonas aeruginosa precipitins determined by means of crossed immunoelectrophoresis. A survey. Acta Pathol Microbiol Scand Suppl 262:1–96. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

