ZNF50 3/ Zpo2 drives aggressive breast cancer progression by down-regulation of GATA3 expression
- PMID: 28258171
- PMCID: PMC5373372
- DOI: 10.1073/pnas.1701690114
ZNF50 3/ Zpo2 drives aggressive breast cancer progression by down-regulation of GATA3 expression
Abstract
The transcription factor GATA3 is the master regulator that drives mammary luminal epithelial cell differentiation and maintains mammary gland homeostasis. Loss of GATA3 is associated with aggressive breast cancer development. We have identified ZNF503/ZEPPO2 zinc-finger elbow-related proline domain protein 2 (ZPO2) as a transcriptional repressor of GATA3 expression and transcriptional activity that induces mammary epithelial cell proliferation and breast cancer development. We show that ZPO2 is recruited to GATA3 promoter in association with ZBTB32 (Repressor of GATA, ROG) and that ZBTB32 is essential for down-regulation of GATA3 via ZPO2. Through this modulation of GATA3 activity, ZPO2 promotes aggressive breast cancer development. Our data provide insight into a mechanism of GATA3 regulation, and identify ZPO2 as a possible candidate gene for future diagnostic and therapeutic strategies.
Keywords: GATA3; ZBTB32; ZNF503/ZPO2; breast cancer; tumor metastasis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
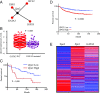






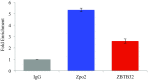
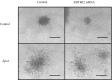
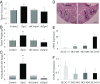


Similar articles
-
The Transcriptional Repressor ZNF503/Zeppo2 Promotes Mammary Epithelial Cell Proliferation and Enhances Cell Invasion.J Biol Chem. 2015 Feb 6;290(6):3803-13. doi: 10.1074/jbc.M114.611202. Epub 2014 Dec 23. J Biol Chem. 2015. PMID: 25538248 Free PMC article.
-
Progesterone receptor activation downregulates GATA3 by transcriptional repression and increased protein turnover promoting breast tumor growth.Breast Cancer Res. 2014 Dec 6;16(6):491. doi: 10.1186/s13058-014-0491-x. Breast Cancer Res. 2014. PMID: 25479686 Free PMC article.
-
GATA3 targets semaphorin 3B in mammary epithelial cells to suppress breast cancer progression and metastasis.Oncogene. 2017 Oct 5;36(40):5567-5575. doi: 10.1038/onc.2017.165. Epub 2017 Jun 5. Oncogene. 2017. PMID: 28581515 Free PMC article.
-
GATA3 in Breast Cancer: Tumor Suppressor or Oncogene?Gene Expr. 2015;16(4):163-8. doi: 10.3727/105221615X14399878166113. Gene Expr. 2015. PMID: 26637396 Free PMC article. Review.
-
GATA3 in development and cancer differentiation: cells GATA have it!J Cell Physiol. 2010 Jan;222(1):42-9. doi: 10.1002/jcp.21943. J Cell Physiol. 2010. PMID: 19798694 Free PMC article. Review.
Cited by
-
MicroRNA-495: a therapeutic and diagnostic tumor marker.J Mol Histol. 2023 Dec;54(6):559-578. doi: 10.1007/s10735-023-10159-0. Epub 2023 Sep 28. J Mol Histol. 2023. PMID: 37759132 Review.
-
A panel of Transcription factors identified by data mining can predict the prognosis of head and neck squamous cell carcinoma.Cancer Cell Int. 2019 Nov 15;19:297. doi: 10.1186/s12935-019-1024-6. eCollection 2019. Cancer Cell Int. 2019. PMID: 31787845 Free PMC article.
-
Znf703 is a novel RA target in the neural plate border.Sci Rep. 2019 Jun 4;9(1):8275. doi: 10.1038/s41598-019-44722-1. Sci Rep. 2019. PMID: 31164691 Free PMC article.
-
Overexpressed GATA3 enhances the sensitivity of colorectal cancer cells to oxaliplatin through regulating MiR-29b.Cancer Cell Int. 2020 Jul 24;20:339. doi: 10.1186/s12935-020-01424-3. eCollection 2020. Cancer Cell Int. 2020. PMID: 32760217 Free PMC article.
-
Expression Profiles and Prognostic Value of FABPs in Colorectal Adenocarcinomas.Biomedicines. 2021 Oct 13;9(10):1460. doi: 10.3390/biomedicines9101460. Biomedicines. 2021. PMID: 34680577 Free PMC article.
References
-
- Asselin-Labat ML, et al. Gata-3 is an essential regulator of mammary gland morphogenesis and luminal cell differentiation. Nat Cell Biol. 2007;9(2):201–209. - PubMed
-
- Bertucci F, et al. Gene expression profiling of primary breast carcinomas using arrays of candidate genes. Hum Mol Genet. 2000;9(20):2981–2991. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

