Downregulation of E Protein Activity Augments an ILC2 Differentiation Program in the Thymus
- PMID: 28258196
- PMCID: PMC5404348
- DOI: 10.4049/jimmunol.1602009
Downregulation of E Protein Activity Augments an ILC2 Differentiation Program in the Thymus
Abstract
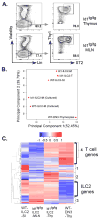
Innate lymphoid cells (ILCs) are important regulators in various immune responses. The current paradigm states that all newly made ILCs originate from common lymphoid progenitors in the bone marrow. Id2, an inhibitor of E protein transcription factors, is indispensable for ILC differentiation. Unexpectedly, we found that ectopically expressing Id1 or deleting two E protein genes in the thymus drastically increased ILC2 counts in the thymus and other organs where ILC2 normally reside. Further evidence suggests a thymic origin of these mutant ILC2s. The mutant mice exhibit augmented spontaneous infiltration of eosinophils and heightened responses to papain in the lung and increased ability to expulse the helminth parasite, Nippostrongylus brasiliensis These results prompt the questions of whether the thymus naturally has the capacity to produce ILC2s and whether E proteins restrain such a potential. The abundance of ILC2s in Id1 transgenic mice also offers a unique opportunity for testing the biological functions of ILC2s.
Copyright © 2017 by The American Association of Immunologists, Inc.
Figures






References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

