Molecular mechanism and structural basis of gain-of-function of STAT1 caused by pathogenic R274Q mutation
- PMID: 28258222
- PMCID: PMC5391754
- DOI: 10.1074/jbc.M116.753848
Molecular mechanism and structural basis of gain-of-function of STAT1 caused by pathogenic R274Q mutation
Abstract
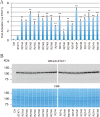
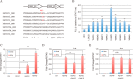
Gain-of-function (GOF) mutations in the STAT1 gene are critical for the onset of chronic mucocutaneous candidiasis (CMC) disease. However, the molecular basis for the gain of STAT1 function remains largely unclear. Here, we investigated the structural features of STAT1 GOF residues to better understand the impact of these pathogenic mutations. We constructed STAT1 alanine mutants of the α3 helix residues of the coiled-coil domain, which are frequently found in CMC pathogenic mutations, and measured their transcriptional activities. Most of the identified GOF residues were located inside the coiled-coil domain stem structure or at the protein surface of the anti-parallel dimer interface. Unlike those, Arg-274 was adjacent to the DNA-binding domain. In addition, Arg-274 was found to functionally interact with Gln-441 in the DNA-binding domain. Because Gln-441 is located at the anti-parallel dimer contact site, Gln-441 reorientation by Arg-274 mutation probably impedes formation of the dimer. Further, the statistical analysis of RNA-seq data with STAT1-deficient epithelial cells and primary T cells from a CMC patient revealed that the R274Q mutation affected gene expression levels of 66 and 76 non-overlapping RefSeq genes, respectively. Because their transcription levels were only slightly modulated by wild-type STAT1, we concluded that the R274Q mutation increased transcriptional activity but did not change dramatically the repertoire of STAT1 targets. Hence, we provide a novel mechanism of STAT1 GOF triggered by a CMC pathogenic mutation.
Keywords: Janus kinase (JAK); genetics; pathogenesis; signal transducers and activators of transcription 1 (STAT1); transcription regulation.
© 2017 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures






References
-
- Shuai K., and Liu B. (2003) Regulation of JAK-STAT signalling in the immune system. Nat. Rev. Immunol. 3, 900–911 - PubMed
-
- Chapgier A., Boisson-Dupuis S., Jouanguy E., Vogt G., Feinberg J., Prochnicka-Chalufour A., Casrouge A., Yang K., Soudais C., Fieschi C., Santos O. F., Bustamante J., Picard C., de Beaucoudrey L., Emile J. F., et al. (2006) Novel STAT1 alleles in otherwise healthy patients with mycobacterial disease. PLoS Genet. 2, e131. - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

