Critical Issues in Mycobiota Analysis
- PMID: 28261162
- PMCID: PMC5306204
- DOI: 10.3389/fmicb.2017.00180
Critical Issues in Mycobiota Analysis
Abstract
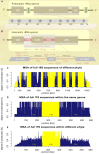
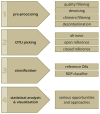
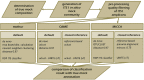
Fungi constitute an important part of the human microbiota and they play a significant role for health and disease development. Advancements made in the culture-independent analysis of microbial communities have broadened our understanding of the mycobiota, however, microbiota analysis tools have been mainly developed for bacteria (e.g., targeting the 16S rRNA gene) and they often fall short if applied to fungal marker-gene based investigations (i.e., internal transcribed spacers, ITS). In the current paper we discuss all major steps of a fungal amplicon analysis starting with DNA extraction from specimens up to bioinformatics analyses of next-generation sequencing data. Specific points are discussed at each step and special emphasis is placed on the bioinformatics challenges emerging during operational taxonomic unit (OTU) picking, a critical step in mycobiota analysis. By using an in silico ITS1 mock community we demonstrate that standard analysis pipelines fall short if used with default settings showing erroneous fungal community representations. We highlight that switching OTU picking to a closed reference approach greatly enhances performance. Finally, recommendations are given on how to perform ITS based mycobiota analysis with the currently available measures.
Keywords: 16S rRNA gene; DNA isolation; OTU picking; formalin-fixed paraffin-embedded tissue (FFPE); internal transcribed spacer (ITS); microbiota; multiple sequence alignment (MSA); mycobiota.
Figures



References
-
- Bazzicalupo A. L., Bálint M., Schmitt I. (2013). Comparison of ITS1 and ITS2 rDNA in 454 sequencing of hyperdiverse fungal communities. Fungal Ecol. 6, 102–109. 10.1016/j.funeco.2012.09.003 - DOI
-
- Bengtsson-Palme J., Ryberg M., Hartmann M., Branco S., Wang Z., Godhe A., et al. (2013). Improved software detection and extraction of ITS1 and ITS2 from ribosomal ITS sequences of fungi and other eukaryotes for analysis of environmental sequencing data. Methods Ecol. Evol. 4, 914–919. 10.1111/2041-210x.12073 - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources

