Common Distribution of gad Operon in Lactobacillus brevis and its GadA Contributes to Efficient GABA Synthesis toward Cytosolic Near-Neutral pH
- PMID: 28261168
- PMCID: PMC5306213
- DOI: 10.3389/fmicb.2017.00206
Common Distribution of gad Operon in Lactobacillus brevis and its GadA Contributes to Efficient GABA Synthesis toward Cytosolic Near-Neutral pH
Abstract
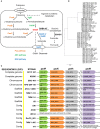
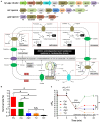
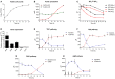
Many strains of lactic acid bacteria (LAB) and bifidobacteria have exhibited strain-specific capacity to produce γ-aminobutyric acid (GABA) via their glutamic acid decarboxylase (GAD) system, which is one of amino acid-dependent acid resistance (AR) systems in bacteria. However, the linkage between bacterial AR and GABA production capacity has not been well established. Meanwhile, limited evidence has been provided to the global diversity of GABA-producing LAB and bifidobacteria, and their mechanisms of efficient GABA synthesis. In this study, genomic survey identified common distribution of gad operon-encoded GAD system in Lactobacillus brevis for its GABA production among varying species of LAB and bifidobacteria. Importantly, among four commonly distributed amino acid-dependent AR systems in Lb. brevis, its GAD system was a major contributor to maintain cytosolic pH homeostasis by consuming protons via GABA synthesis. This highlights that Lb. brevis applies GAD system as the main strategy against extracellular and intracellular acidification demonstrating its high capacity of GABA production. In addition, the abundant GadA retained its activity toward near-neutral pH (pH 5.5-6.5) of cytosolic acidity thus contributing to efficient GABA synthesis in Lb. brevis. This is the first global report illustrating species-specific characteristic and mechanism of efficient GABA synthesis in Lb. brevis.
Keywords: Lactobacillus brevis; acid resistance; genomic survey; glutamic acid decarboxylase; γ-aminobutyric acid (GABA).
Figures

Similar articles
-
Restoration of GABA production machinery in Lactobacillus brevis by accessible carbohydrates, anaerobiosis and early acidification.Food Microbiol. 2018 Feb;69:151-158. doi: 10.1016/j.fm.2017.08.006. Epub 2017 Aug 16. Food Microbiol. 2018. PMID: 28941896
-
Exploring the contributions of two glutamate decarboxylase isozymes in Lactobacillus brevis to acid resistance and γ-aminobutyric acid production.Microb Cell Fact. 2018 Nov 19;17(1):180. doi: 10.1186/s12934-018-1029-1. Microb Cell Fact. 2018. PMID: 30454056 Free PMC article.
-
High γ-aminobutyric acid production from lactic acid bacteria: Emphasis on Lactobacillus brevis as a functional dairy starter.Crit Rev Food Sci Nutr. 2017 Nov 22;57(17):3661-3672. doi: 10.1080/10408398.2016.1147418. Crit Rev Food Sci Nutr. 2017. PMID: 26980301 Review.
-
Comparative Peptidomic and Metatranscriptomic Analyses Reveal Improved Gamma-Amino Butyric Acid Production Machinery in Levilactobacillus brevis Strain NPS-QW 145 Cocultured with Streptococcus thermophilus Strain ASCC1275 during Milk Fermentation.Appl Environ Microbiol. 2020 Dec 17;87(1):e01985-20. doi: 10.1128/AEM.01985-20. Print 2020 Dec 17. Appl Environ Microbiol. 2020. PMID: 33067198 Free PMC article.
-
Glutamate Decarboxylase from Lactic Acid Bacteria-A Key Enzyme in GABA Synthesis.Microorganisms. 2020 Dec 3;8(12):1923. doi: 10.3390/microorganisms8121923. Microorganisms. 2020. PMID: 33287375 Free PMC article. Review.
Cited by
-
Metagenomics-based inference of microbial metabolism towards neuroactive amino acids and the response to antibiotics in piglet colon.Amino Acids. 2023 Oct;55(10):1333-1347. doi: 10.1007/s00726-023-03311-3. Epub 2023 Aug 15. Amino Acids. 2023. PMID: 37581868
-
Functional Properties and Sustainability Improvement of Sourdough Bread by Lactic Acid Bacteria.Microorganisms. 2020 Nov 30;8(12):1895. doi: 10.3390/microorganisms8121895. Microorganisms. 2020. PMID: 33265943 Free PMC article.
-
The role of GABA in type 1 diabetes.Front Endocrinol (Lausanne). 2024 Nov 15;15:1453396. doi: 10.3389/fendo.2024.1453396. eCollection 2024. Front Endocrinol (Lausanne). 2024. PMID: 39619323 Free PMC article. Review.
-
GABA synthesizing lactic acid bacteria and genomic analysis of Levilactobacillus brevis LAB6.3 Biotech. 2024 Mar;14(3):62. doi: 10.1007/s13205-024-03918-7. Epub 2024 Feb 7. 3 Biotech. 2024. PMID: 38344283 Free PMC article.
-
Metabolic engineering of microorganisms for the production of multifunctional non-protein amino acids: γ-aminobutyric acid and δ-aminolevulinic acid.Microb Biotechnol. 2021 Nov;14(6):2279-2290. doi: 10.1111/1751-7915.13783. Epub 2021 Mar 6. Microb Biotechnol. 2021. PMID: 33675575 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

