Possible Role of MADS AFFECTING FLOWERING 3 and B-BOX DOMAIN PROTEIN 19 in Flowering Time Regulation of Arabidopsis Mutants with Defects in Nonsense-Mediated mRNA Decay
- PMID: 28261246
- PMCID: PMC5306368
- DOI: 10.3389/fpls.2017.00191
Possible Role of MADS AFFECTING FLOWERING 3 and B-BOX DOMAIN PROTEIN 19 in Flowering Time Regulation of Arabidopsis Mutants with Defects in Nonsense-Mediated mRNA Decay
Abstract
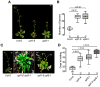
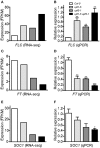
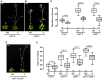
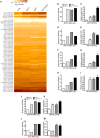
Eukaryotic cells use nonsense-mediated mRNA decay (NMD) to clear aberrant mRNAs from the cell, thus preventing the accumulation of truncated proteins. In Arabidopsis, two UP-Frameshift (UPF) proteins, UPF1 and UPF3, play a critical role in NMD. Although deficiency of UPF1 and UPF3 leads to various developmental defects, little is known about the mechanism underlying the regulation of flowering time by NMD. Here, we showed that the upf1-5 and upf3-1 mutants had a late-flowering phenotype under long-day conditions and the upf1-5 upf3-1 double mutants had an additive effect in delaying flowering time. RNA sequencing of the upf mutants revealed that UPF3 exerted a stronger effect than UPF1 in the UPF-mediated regulation of flowering time. Among genes known to regulate flowering time, FLOWERING LOCUS C (FLC) mRNA levels increased (up to 8-fold) in upf mutants, as confirmed by qPCR. The upf1-5, upf3-1, and upf1-5 upf3-1 mutants responded to vernalization, suggesting a role of FLC in delayed flowering of upf mutants. Consistent with the high FLC transcript levels and delayed flowering in upf mutants, levels of FLOWERING LOCUS T (FT) and SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 (SOC1) mRNAs were reduced in the upf mutants. However, RNA-seq did not identify an aberrant FLC transcript containing a premature termination codon (PTC), suggesting that FLC is not a direct target in the regulation of flowering time by NMD. Among flowering time regulators that act in an FLC-dependent manner, we found that MAF3, NF-YA2, NF-YA5, and TAF14 showed increased transcript levels in upf mutants. We also found that BBX19 and ATC, which act in an FLC-independent manner, showed increased transcript levels in upf mutants. An aberrant transcript containing a PTC was identified from MAF3 and BBX19 and the levels of the aberrant transcripts increased in upf mutants. Taking these results together, we propose that the late-flowering phenotype of upf mutants is mediated by at least two different pathways, namely, by MAF3 in an FLC-dependent manner and by BBX19 in an FLC-independent manner.
Keywords: Arabidopsis; NMD; UPF1; UPF3; flowering.
Figures



Similar articles
-
Nonsense-mediated mRNA decay modulates Arabidopsis flowering time via the SET DOMAIN GROUP 40-FLOWERING LOCUS C module.J Exp Bot. 2021 Oct 26;72(20):7049-7066. doi: 10.1093/jxb/erab331. J Exp Bot. 2021. PMID: 34270724
-
UPF1 is required for nonsense-mediated mRNA decay (NMD) and RNAi in Arabidopsis.Plant J. 2006 Aug;47(3):480-9. doi: 10.1111/j.1365-313X.2006.02802.x. Epub 2006 Jun 30. Plant J. 2006. PMID: 16813578
-
Nonsense-mediated mRNA decay factors, UPF1 and UPF3, contribute to plant defense.Plant Cell Physiol. 2011 Dec;52(12):2147-56. doi: 10.1093/pcp/pcr144. Epub 2011 Oct 24. Plant Cell Physiol. 2011. PMID: 22025558
-
NMD: At the crossroads between translation termination and ribosome recycling.Biochimie. 2015 Jul;114:2-9. doi: 10.1016/j.biochi.2014.10.027. Epub 2014 Nov 13. Biochimie. 2015. PMID: 25446649 Free PMC article. Review.
-
Nonsense-Mediated mRNA Decay: Degradation of Defective Transcripts Is Only Part of the Story.Annu Rev Genet. 2015;49:339-66. doi: 10.1146/annurev-genet-112414-054639. Epub 2015 Oct 2. Annu Rev Genet. 2015. PMID: 26436458 Free PMC article. Review.
Cited by
-
Mining and expression analysis of candidate genes involved in regulating the chilling requirement fulfillment of Paeonia lactiflora 'Hang Baishao'.BMC Plant Biol. 2017 Dec 22;17(1):262. doi: 10.1186/s12870-017-1205-1. BMC Plant Biol. 2017. PMID: 29273002 Free PMC article.
-
Polyamines as Quality Control Metabolites Operating at the Post-Transcriptional Level.Plants (Basel). 2019 Apr 24;8(4):109. doi: 10.3390/plants8040109. Plants (Basel). 2019. PMID: 31022874 Free PMC article. Review.
-
BcMAF2 activates BcTEM1 and represses flowering in Pak-choi (Brassica rapa ssp. chinensis).Plant Mol Biol. 2019 May;100(1-2):19-32. doi: 10.1007/s11103-019-00867-1. Epub 2019 Apr 18. Plant Mol Biol. 2019. PMID: 31001712
-
Molecular Characterization and Expression Profile of PaCOL1, a CONSTANS-like Gene in Phalaenopsis Orchid.Plants (Basel). 2020 Jan 4;9(1):68. doi: 10.3390/plants9010068. Plants (Basel). 2020. PMID: 31947959 Free PMC article.
-
The Molecular Basis of Kale Domestication: Transcriptional Profiling of Developing Leaves Provides New Insights Into the Evolution of a Brassica oleracea Vegetative Morphotype.Front Plant Sci. 2021 Mar 5;12:637115. doi: 10.3389/fpls.2021.637115. eCollection 2021. Front Plant Sci. 2021. PMID: 33747016 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

