Transcript Profiling Reveals the Presence of Abiotic Stress and Developmental Stage Specific Ascorbate Oxidase Genes in Plants
- PMID: 28261251
- PMCID: PMC5314155
- DOI: 10.3389/fpls.2017.00198
Transcript Profiling Reveals the Presence of Abiotic Stress and Developmental Stage Specific Ascorbate Oxidase Genes in Plants
Abstract
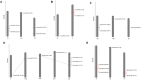
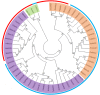
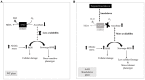
Abiotic stress and climate change is the major concern for plant growth and crop yield. Abiotic stresses lead to enhanced accumulation of reactive oxygen species (ROS) consequently resulting in cellular damage and major losses in crop yield. One of the major scavengers of ROS is ascorbate (AA) which acts as first line of defense against external oxidants. An enzyme named ascorbate oxidase (AAO) is known to oxidize AA and deleteriously affect the plant system in response to stress. Genome-wide analysis of AAO gene family has led to the identification of five, three, seven, four, and six AAO genes in Oryza sativa, Arabidopsis, Glycine max, Zea mays, and Sorghum bicolor genomes, respectively. Expression profiling of these genes was carried out in response to various abiotic stresses and during various stages of vegetative and reproductive development using publicly available microarray database. Expression analysis in Oryza sativa revealed tissue specific expression of AAO genes wherein few members were exclusively expressed in either root or shoot. These genes were found to be regulated by both developmental cues as well as diverse stress conditions. The qRT-PCR analysis in response to salinity and drought stress in rice shoots revealed OsAAO2 to be the most stress responsive gene. On the other hand, OsAAO3 and OsAAO4 genes showed enhanced expression in roots under salinity/drought stresses. This study provides lead about important stress responsive AAO genes in various crop plants, which could be used to engineer climate resilient crop plants.
Keywords: abiotic stress; ascorbate oxidase; genome-wide analysis; qRT-PCR; reactive oxygen species (ROS).
Figures




Similar articles
-
Characteristic of the Ascorbate Oxidase Gene Family in Beta vulgaris and Analysis of the Role of AAO in Response to Salinity and Drought in Beet.Int J Mol Sci. 2022 Oct 23;23(21):12773. doi: 10.3390/ijms232112773. Int J Mol Sci. 2022. PMID: 36361565 Free PMC article.
-
Cross-species multiple environmental stress responses: An integrated approach to identify candidate genes for multiple stress tolerance in sorghum (Sorghum bicolor (L.) Moench) and related model species.PLoS One. 2018 Mar 28;13(3):e0192678. doi: 10.1371/journal.pone.0192678. eCollection 2018. PLoS One. 2018. PMID: 29590108 Free PMC article.
-
Genome-wide analysis and expression profiling of glyoxalase gene families in soybean (Glycine max) indicate their development and abiotic stress specific response.BMC Plant Biol. 2016 Apr 16;16:87. doi: 10.1186/s12870-016-0773-9. BMC Plant Biol. 2016. PMID: 27083416 Free PMC article.
-
Reactive Oxygen Species and Antioxidant Defense in Plants under Abiotic Stress: Revisiting the Crucial Role of a Universal Defense Regulator.Antioxidants (Basel). 2020 Jul 29;9(8):681. doi: 10.3390/antiox9080681. Antioxidants (Basel). 2020. PMID: 32751256 Free PMC article. Review.
-
An Overview of Abiotic Stress in Cereal Crops: Negative Impacts, Regulation, Biotechnology and Integrated Omics.Plants (Basel). 2021 Jul 19;10(7):1472. doi: 10.3390/plants10071472. Plants (Basel). 2021. PMID: 34371676 Free PMC article. Review.
Cited by
-
24-Epibrassinolide (EBR) Confers Tolerance against NaCl Stress in Soybean Plants by Up-Regulating Antioxidant System, Ascorbate-Glutathione Cycle, and Glyoxalase System.Biomolecules. 2019 Oct 23;9(11):640. doi: 10.3390/biom9110640. Biomolecules. 2019. PMID: 31652728 Free PMC article.
-
Comparative Proteomics Identified Proteins in Mung Bean Sprouts Under Different Concentrations of Urea.Molecules. 2025 Jul 29;30(15):3176. doi: 10.3390/molecules30153176. Molecules. 2025. PMID: 40807351 Free PMC article.
-
C-terminally encoded peptides (CEPs) are potential mediators of abiotic stress response in plants.Physiol Mol Biol Plants. 2020 Oct;26(10):2019-2033. doi: 10.1007/s12298-020-00881-4. Epub 2020 Sep 21. Physiol Mol Biol Plants. 2020. PMID: 33088046 Free PMC article.
-
Genome-Wide Identification and Expression Analysis of the Ascorbate Oxidase Gene Family in Gossypium hirsutum Reveals the Critical Role of GhAO1A in Delaying Dark-Induced Leaf Senescence.Int J Mol Sci. 2019 Dec 6;20(24):6167. doi: 10.3390/ijms20246167. Int J Mol Sci. 2019. PMID: 31817730 Free PMC article.
-
Gene Profiling of the Ascorbate Oxidase Family Genes under Osmotic and Cold Stress Reveals the Role of AnAO5 in Cold Adaptation in Ammopiptanthus nanus.Plants (Basel). 2023 Feb 3;12(3):677. doi: 10.3390/plants12030677. Plants (Basel). 2023. PMID: 36771760 Free PMC article.
References
-
- Balestrini R., Ott T., Güther M., Bonfante P., Udvardi M. K., De Tullio M. C. (2012). Ascorbate oxidase: the unexpected involvement of a “wasteful enzyme” in the symbioses with nitrogen-fixing bacteria and arbuscular mycorrhizal fungi. Plant Physiol. Biochem. 59 71–79. 10.1016/j.plaphy.2012.07.006 - DOI - PubMed
-
- Barnes J., Zheng Y., Lyons T. (2002). “Plant resistance to ozone: the role of ascorbate,” in Air pollution and Plant Biotechnology eds Omasa K., Saji H., Youssefian S., Kondo N. (Tokyo: Springer; ) 235–252.
LinkOut - more resources
Full Text Sources
Other Literature Sources

