In silico Comparison of 19 Porphyromonas gingivalis Strains in Genomics, Phylogenetics, Phylogenomics and Functional Genomics
- PMID: 28261563
- PMCID: PMC5306136
- DOI: 10.3389/fcimb.2017.00028
In silico Comparison of 19 Porphyromonas gingivalis Strains in Genomics, Phylogenetics, Phylogenomics and Functional Genomics
Abstract
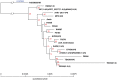
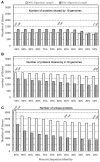
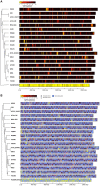
Currently, genome sequences of a total of 19 Porphyromonas gingivalis strains are available, including eight completed genomes (strains W83, ATCC 33277, TDC60, HG66, A7436, AJW4, 381, and A7A1-28) and 11 high-coverage draft sequences (JCVI SC001, F0185, F0566, F0568, F0569, F0570, SJD2, W4087, W50, Ando, and MP4-504) that are assembled into fewer than 300 contigs. The objective was to compare these genomes at both nucleotide and protein sequence levels in order to understand their phylogenetic and functional relatedness. Four copies of 16S rRNA gene sequences were identified in each of the eight complete genomes and one in the other 11 unfinished genomes. These 43 16S rRNA sequences represent only 24 unique sequences and the derived phylogenetic tree suggests a possible evolutionary history for these strains. Phylogenomic comparison based on shared proteins and whole genome nucleotide sequences consistently showed two groups with closely related members: one consisted of ATCC 33277, 381, and HG66, another of W83, W50, and A7436. At least 1,037 core/shared proteins were identified in the 19 P. gingivalis genomes based on the most stringent detecting parameters. Comparative functional genomics based on genome-wide comparisons between NCBI and RAST annotations, as well as additional approaches, revealed functions that are unique or missing in individual P. gingivalis strains, or species-specific in all P. gingivalis strains, when compared to a neighboring species P. asaccharolytica. All the comparative results of this study are available online for download at ftp://www.homd.org/publication_data/20160425/.
Keywords: Porphyromonas gingivalis; comparative genomics; phylogenetics; phylogenomics.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

