Pseudomonas aeruginosa Lifestyle: A Paradigm for Adaptation, Survival, and Persistence
- PMID: 28261568
- PMCID: PMC5310132
- DOI: 10.3389/fcimb.2017.00039
Pseudomonas aeruginosa Lifestyle: A Paradigm for Adaptation, Survival, and Persistence
Abstract
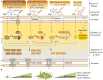
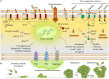
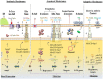
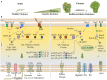
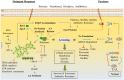
Pseudomonas aeruginosa is an opportunistic pathogen affecting immunocompromised patients. It is known as the leading cause of morbidity and mortality in cystic fibrosis (CF) patients and as one of the leading causes of nosocomial infections. Due to a range of mechanisms for adaptation, survival and resistance to multiple classes of antibiotics, infections by P. aeruginosa strains can be life-threatening and it is emerging worldwide as public health threat. This review highlights the diversity of mechanisms by which P. aeruginosa promotes its survival and persistence in various environments and particularly at different stages of pathogenesis. We will review the importance and complexity of regulatory networks and genotypic-phenotypic variations known as adaptive radiation by which P. aeruginosa adjusts physiological processes for adaptation and survival in response to environmental cues and stresses. Accordingly, we will review the central regulatory role of quorum sensing and signaling systems by nucleotide-based second messengers resulting in different lifestyles of P. aeruginosa. Furthermore, various regulatory proteins will be discussed which form a plethora of controlling systems acting at transcriptional level for timely expression of genes enabling rapid responses to external stimuli and unfavorable conditions. Antibiotic resistance is a natural trait for P. aeruginosa and multiple mechanisms underlying different forms of antibiotic resistance will be discussed here. The importance of each mechanism in conferring resistance to various antipseudomonal antibiotics and their prevalence in clinical strains will be described. The underlying principles for acquiring resistance leading pan-drug resistant strains will be summarized. A future outlook emphasizes the need for collaborative international multidisciplinary efforts to translate current knowledge into strategies to prevent and treat P. aeruginosa infections while reducing the rate of antibiotic resistance and avoiding the spreading of resistant strains.
Keywords: Pseudomonas aeruginosa; antibiotic resistance; biofilm; persistence; virulence.
Figures
References
-
- Aendekerk S., Diggle S. P., Song Z., Høiby N., Cornelis P., Williams P., et al. . (2005). The MexGHI-OpmD multidrug efflux pump controls growth, antibiotic susceptibility and virulence in Pseudomonas aeruginosa via 4-quinolone-dependent cell-to-cell communication. Microbiology 151(Pt 4), 1113–1125. 10.1099/mic.0.27631-0 - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

