Development of a Proximity Labeling System to Map the Chlamydia trachomatis Inclusion Membrane
- PMID: 28261569
- PMCID: PMC5309262
- DOI: 10.3389/fcimb.2017.00040
Development of a Proximity Labeling System to Map the Chlamydia trachomatis Inclusion Membrane
Abstract
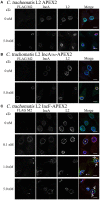
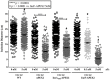
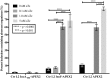
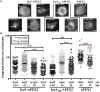
Chlamydia grows within a membrane-bound vacuole termed an inclusion. The cellular processes that support the biogenesis and integrity of this pathogen-specified parasitic organelle are not understood. Chlamydia secretes integral membrane proteins called Incs that insert into the chlamydial inclusion membrane (IM). Incs contain at least two hydrophobic transmembrane domains flanked by termini, which vary in size and are exposed to the host cytosol. In addition, Incs are temporally expressed during the chlamydial developmental cycle. Data examining Inc function are limited because of (i) the difficulty in working with hydrophobic proteins and (ii) the inherent fragility of the IM. We hypothesize that Incs function collaboratively to maintain the integrity of the chlamydial inclusion with small Incs organizing the IM and larger Incs interfacing with host cell machinery. To study this hypothesis, we have adapted a proximity-labeling strategy using APEX2, a mutant soybean ascorbate peroxidase that biotinylates interacting and proximal proteins within minutes in the presence of H2O2 and its exogenous substrate, biotin-phenol. We successfully expressed, from an inducible background, APEX2 alone, or fusion proteins of IncATM (TM = transmembrane domain only), IncA, and IncF with APEX2 in Chlamydia trachomatis serovar L2. IncF-APEX2, IncA TM -APEX2, and IncA-APEX2 localized to the IM whereas APEX2, lacking a secretion signal, remained associated with the bacteria. We determined the impact of overexpression on inclusion diameter, plasmid stability, and Golgi-derived sphingomyelin acquisition. While there was an overall impact of inducing construct expression, IncF-APEX2 overexpression most negatively impacted these measurements. Importantly, Inc-APEX2 expression in the presence of biotin-phenol resulted in biotinylation of the IM. These data suggest that Inc expression is regulated to control optimal IM biogenesis. We subsequently defined lysis conditions that solubilized known Incs and were compatible with pulldown conditions. Importantly, we have created powerful tools to allow direct examination of the dynamic composition of the IM, which will provide novel insights into key interactions that promote chlamydial growth and development within the inclusion.
Keywords: Chlamydia trachomatis; Inc; inclusion membrane; proximity labeling.
Figures
Similar articles
-
Proximity Labeling To Map Host-Pathogen Interactions at the Membrane of a Bacterium-Containing Vacuole in Chlamydia trachomatis-Infected Human Cells.Infect Immun. 2019 Oct 18;87(11):e00537-19. doi: 10.1128/IAI.00537-19. Print 2019 Nov. Infect Immun. 2019. PMID: 31405957 Free PMC article.
-
Proximity Labeling of the Chlamydia trachomatis Inclusion Membrane.Methods Mol Biol. 2019;2042:245-278. doi: 10.1007/978-1-4939-9694-0_17. Methods Mol Biol. 2019. PMID: 31385281
-
A meta-analysis of affinity purification-mass spectrometry experimental systems used to identify eukaryotic and chlamydial proteins at the Chlamydia trachomatis inclusion membrane.J Proteomics. 2020 Feb 10;212:103595. doi: 10.1016/j.jprot.2019.103595. Epub 2019 Nov 21. J Proteomics. 2020. PMID: 31760040 Free PMC article.
-
Safe haven under constant attack-The Chlamydia-containing vacuole.Cell Microbiol. 2018 Oct;20(10):e12940. doi: 10.1111/cmi.12940. Epub 2018 Sep 4. Cell Microbiol. 2018. PMID: 30101516 Review.
-
Chlamydia trachomatis and its interaction with the cellular retromer.Int J Med Microbiol. 2018 Jan;308(1):197-205. doi: 10.1016/j.ijmm.2017.10.006. Epub 2017 Oct 26. Int J Med Microbiol. 2018. PMID: 29122514 Review.
Cited by
-
Initial Characterization of the Two ClpP Paralogs of Chlamydia trachomatis Suggests Unique Functionality for Each.J Bacteriol. 2018 Dec 20;201(2):e00635-18. doi: 10.1128/JB.00635-18. Print 2019 Jan 15. J Bacteriol. 2018. PMID: 30396899 Free PMC article.
-
Proximity-dependent proteomics of the Chlamydia trachomatis inclusion membrane reveals functional interactions with endoplasmic reticulum exit sites.PLoS Pathog. 2019 Apr 3;15(4):e1007698. doi: 10.1371/journal.ppat.1007698. eCollection 2019 Apr. PLoS Pathog. 2019. PMID: 30943267 Free PMC article.
-
Establishing the intracellular niche of obligate intracellular vacuolar pathogens.Front Cell Infect Microbiol. 2023 Aug 14;13:1206037. doi: 10.3389/fcimb.2023.1206037. eCollection 2023. Front Cell Infect Microbiol. 2023. PMID: 37645379 Free PMC article. Review.
-
Type III Secretion in Chlamydia.Microbiol Mol Biol Rev. 2023 Sep 26;87(3):e0003423. doi: 10.1128/mmbr.00034-23. Epub 2023 Jun 26. Microbiol Mol Biol Rev. 2023. PMID: 37358451 Free PMC article. Review.
-
The growing repertoire of genetic tools for dissecting chlamydial pathogenesis.Pathog Dis. 2021 May 11;79(5):ftab025. doi: 10.1093/femspd/ftab025. Pathog Dis. 2021. PMID: 33930127 Free PMC article.
References
-
- Aeberhard L., Banhart S., Fischer M., Jehmlich N., Rose L., Koch S., et al. . (2015). The Proteome of the isolated Chlamydia trachomatis containing vacuole reveals a complex trafficking platform enriched for retromer components. PLoS Pathog. 11:e1004883. 10.1371/journal.ppat.1004883 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

