Divergence of annual and perennial species in the Brassicaceae and the contribution of cis-acting variation at FLC orthologues
- PMID: 28261921
- PMCID: PMC5485006
- DOI: 10.1111/mec.14084
Divergence of annual and perennial species in the Brassicaceae and the contribution of cis-acting variation at FLC orthologues
Abstract
Variation in life history contributes to reproductive success in different environments. Divergence of annual and perennial angiosperm species is an extreme example that has occurred frequently. Perennials survive for several years and restrict the duration of reproduction by cycling between vegetative growth and flowering, whereas annuals live for 1 year and flower once. We used the tribe Arabideae (Brassicaceae) to study the divergence of seasonal flowering behaviour among annual and perennial species. In perennial Brassicaceae, orthologues of FLOWERING LOCUS C (FLC), a floral inhibitor in Arabidopsis thaliana, are repressed by winter cold and reactivated in spring conferring seasonal flowering patterns, whereas in annuals, they are stably repressed by cold. We isolated FLC orthologues from three annual and two perennial Arabis species and found that the duplicated structure of the A. alpina locus is not required for perenniality. The expression patterns of the genes differed between annuals and perennials, as observed among Arabidopsis species, suggesting a broad relevance of these patterns within the Brassicaceae. Also analysis of plants derived from an interspecies cross of A. alpina and annual A. montbretiana demonstrated that cis-regulatory changes in FLC orthologues contribute to their different transcriptional patterns. Sequence comparisons of FLC orthologues from annuals and perennials in the tribes Arabideae and Camelineae identified two regulatory regions in the first intron whose sequence variation correlates with divergence of the annual and perennial expression patterns. Thus, we propose that related cis-acting changes in FLC orthologues occur independently in different tribes of the Brassicaceae during life history evolution.
Keywords: Arabis; FLC; PEP1; annual; flowering time; perennial.
© 2017 The Authors. Molecular Ecology Published by John Wiley & Sons Ltd.
Figures


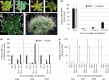
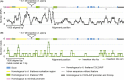


Comment in
-
Variation in gene regulation underlying annual and perennial flowering in Arabideae species.Mol Ecol. 2017 Jul;26(13):3324-3326. doi: 10.1111/mec.14171. Mol Ecol. 2017. PMID: 28632342
Similar articles
-
PEP1 regulates perennial flowering in Arabis alpina.Nature. 2009 May 21;459(7245):423-7. doi: 10.1038/nature07988. Epub 2009 Apr 15. Nature. 2009. PMID: 19369938
-
Variation in gene regulation underlying annual and perennial flowering in Arabideae species.Mol Ecol. 2017 Jul;26(13):3324-3326. doi: 10.1111/mec.14171. Mol Ecol. 2017. PMID: 28632342
-
The Diverse Roles of FLOWERING LOCUS C in Annual and Perennial Brassicaceae Species.Front Plant Sci. 2021 Feb 15;12:627258. doi: 10.3389/fpls.2021.627258. eCollection 2021. Front Plant Sci. 2021. PMID: 33679840 Free PMC article. Review.
-
PEP1 of Arabis alpina is encoded by two overlapping genes that contribute to natural genetic variation in perennial flowering.PLoS Genet. 2012;8(12):e1003130. doi: 10.1371/journal.pgen.1003130. Epub 2012 Dec 20. PLoS Genet. 2012. PMID: 23284298 Free PMC article.
-
Comparative analysis of flowering in annual and perennial plants.Curr Top Dev Biol. 2010;91:323-48. doi: 10.1016/S0070-2153(10)91011-9. Curr Top Dev Biol. 2010. PMID: 20705187 Review.
Cited by
-
Arabis alpina: A perennial model plant for ecological genomics and life-history evolution.Mol Ecol Resour. 2022 Feb;22(2):468-486. doi: 10.1111/1755-0998.13490. Epub 2021 Sep 7. Mol Ecol Resour. 2022. PMID: 34415668 Free PMC article. Review.
-
All roads lead to Rome: QTL analysis for vernalization requirement and dissection of allelic variation uncovered unexpected diversity of FLC loci in Camelina sativa.Front Plant Sci. 2025 Jul 25;16:1639872. doi: 10.3389/fpls.2025.1639872. eCollection 2025. Front Plant Sci. 2025. PMID: 40786938 Free PMC article.
-
Extended Vernalization Regulates Inflorescence Fate in Arabis alpina by Stably Silencing PERPETUAL FLOWERING1.Plant Physiol. 2018 Apr;176(4):2819-2833. doi: 10.1104/pp.17.01754. Epub 2018 Feb 21. Plant Physiol. 2018. PMID: 29467177 Free PMC article.
-
Genetic encoding of complex traits.J Exp Bot. 2021 Jan 20;72(1):1-3. doi: 10.1093/jxb/eraa498. J Exp Bot. 2021. PMID: 33471904 Free PMC article. No abstract available.
-
Beyond a reference genome: pangenomes and population genomics of underutilized and orphan crops for future food and nutrition security.New Phytol. 2022 Jun;234(5):1583-1597. doi: 10.1111/nph.18021. Epub 2022 Mar 22. New Phytol. 2022. PMID: 35318683 Free PMC article. Review.
References
-
- Albani M, Coupland G (2010) Comparative analysis of flowering in annual and perennial plants. Current Topics in Developmental Biology, 91, 323–348. - PubMed
-
- Albani M, Castaings L, Wötzel S et al (2012) PEP1 of Arabis alpina is encoded by two overlapping genes that contribute to natural genetic variation in perennial flowering. PLoS Genetics, 8, e1003130. doi: 10.1371/journal.pgen.1003130. - DOI - PMC - PubMed
-
- Alonso‐Blanco C, Méndez‐Vigo B (2014) Genetic architecture of naturally occurring quantitative traits in plants: an updated synthesis. Current Opinion in Plant Biology, 18, 37–43. - PubMed
-
- Al‐Shehbaz IA, German DA, Karl R, Jordon‐Thaden I, Koch MA (2011) Nomenclatural adjustments in the tribe Arabideae (Brassicaceae). Plant Diversity and Evolution, 129, 71–76.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

