Notch Inhibition Enhances Cardiac Reprogramming by Increasing MEF2C Transcriptional Activity
- PMID: 28262548
- PMCID: PMC5355682
- DOI: 10.1016/j.stemcr.2017.01.025
Notch Inhibition Enhances Cardiac Reprogramming by Increasing MEF2C Transcriptional Activity
Abstract
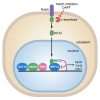
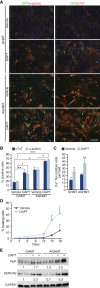
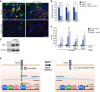
Conversion of fibroblasts into functional cardiomyocytes represents a potential means of restoring cardiac function after myocardial infarction, but so far this process remains inefficient and little is known about its molecular mechanisms. Here we show that DAPT, a classical Notch inhibitor, enhances the conversion of mouse fibroblasts into induced cardiac-like myocytes by the transcription factors GATA4, HAND2, MEF2C, and TBX5. DAPT cooperates with AKT kinase to further augment this process, resulting in up to 70% conversion efficiency. Moreover, DAPT promotes the acquisition of specific cardiomyocyte features, substantially increasing calcium flux, sarcomere structure, and the number of spontaneously beating cells. Transcriptome analysis shows that DAPT induces genetic programs related to muscle development, differentiation, and excitation-contraction coupling. Mechanistically, DAPT increases binding of the transcription factor MEF2C to the promoter regions of cardiac structural genes. These findings provide mechanistic insights into the reprogramming process and may have important implications for cardiac regeneration therapies.
Keywords: DAPT; Notch signaling; cardiomyocytes; cell-fate conversion; direct cellular reprogramming; heart regeneration; regenerative medicine; transdifferentiation.
Copyright © 2017 The Authors. Published by Elsevier Inc. All rights reserved.
Figures



References
-
- Andersson E.R., Sandberg R., Lendahl U. Notch signaling: simplicity in design, versatility in function. Development. 2011;138:3593–3612. - PubMed
-
- Batty J.A., Lima J.A., Jr., Kunadian V. Direct cellular reprogramming for cardiac repair and regeneration. Eur. J. Heart Fail. 2016;18:145–156. - PubMed
-
- Beltrami A.P., Urbanek K., Kajstura J., Yan S.M., Finato N., Bussani R., Nadal-Ginard B., Silvestri F., Leri A., Beltrami C.A. Evidence that human cardiac myocytes divide after myocardial infarction. N. Engl. J. Med. 2001;344:1750–1757. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

