miRNA engineering of CHO cells facilitates production of difficult-to-express proteins and increases success in cell line development
- PMID: 28262952
- PMCID: PMC6084326
- DOI: 10.1002/bit.26280
miRNA engineering of CHO cells facilitates production of difficult-to-express proteins and increases success in cell line development
Abstract
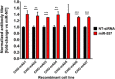
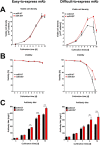
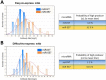
In recent years, coherent with growing biologics portfolios also the number of complex and thus difficult-to-express (DTE) therapeutic proteins has increased considerably. DTE proteins challenge bioprocess development and can include various therapeutic protein formats such as monoclonal antibodies (mAbs), multi-specific affinity scaffolds (e.g., bispecific antibodies), cytokines, or fusion proteins. Hence, the availability of robust and versatile Chinese hamster ovary (CHO) host cell factories is fundamental for high-yielding bioprocesses. MicroRNAs (miRNAs) have emerged as potent cell engineering tools to improve process performance of CHO manufacturing cell lines. However, there has not been any report demonstrating the impact of beneficial miRNAs on industrial cell line development (CLD) yet. To address this question, we established novel CHO host cells constitutively expressing a pro-productive miRNA: miR-557. Novel host cells were tested in two independent CLD campaigns using two different mAb candidates including a normal as well as a DTE antibody. Presence of miR-557 significantly enhanced each process step during CLD in a product independent manner. Stable expression of miR-557 increased the probability to identify high-producing cell clones. Furthermore, production cell lines derived from miR-557 expressing host cells exhibited significantly increased final product yields in fed-batch cultivation processes without compromising product quality. Strikingly, cells co-expressing miR-557 and a DTE antibody achieved a twofold increase in product titer compared to clones co-expressing a negative control miRNA. Thus, host cell engineering using miRNAs represents a promising tool to overcome limitations in industrial CLD especially with regard to DTE proteins. Biotechnol. Bioeng. 2017;114: 1495-1510. © 2017 Wiley Periodicals, Inc.
Keywords: Chinese hamster ovary (CHO) cells; cell engineering; difficult-to-express proteins; miR-557; microRNA; monoclonal antibody.
© 2017 Wiley Periodicals, Inc.
Figures






References
-
- Barron N, Kumar N, Sanchez N, Doolan P, Clarke C, Meleady P, O'Sullivan F, Clynes M. 2011a. Engineering CHO cell growth and recombinant protein productivity by overexpression of miR‐7. J Biotechnol 151(2):204–211. - PubMed
-
- Barron N, Sanchez N, Kelly P, Clynes M. 2011b. MicroRNAs: Tiny targets for engineering CHO cell phenotypes? Biotechnol Lett 33(1):11–21. - PubMed
-
- Bentley KJ, Gewert R, Harris WJ. 1998. Differential efficiency of expression of humanized antibodies in transient transfected mammalian cells. Hybridoma 17(6):559–567. - PubMed
-
- Bertschinger M, Backliwal G, Schertenleib A, Jordan M, Hacker DL, Wurm FM. 2006. Disassembly of polyethylenimine‐DNA particles in vitro: Implications for polyethylenimine‐mediated DNA delivery. J Control Release 116(1):96–104. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

