Uncoupling apical constriction from tissue invagination
- PMID: 28263180
- PMCID: PMC5338918
- DOI: 10.7554/eLife.22235
Uncoupling apical constriction from tissue invagination
Abstract
Apical constriction is a widely utilized cell shape change linked to folding, bending and invagination of polarized epithelia. It remains unclear how apical constriction is regulated spatiotemporally during tissue invagination and how this cellular process contributes to tube formation in different developmental contexts. Using Drosophila salivary gland (SG) invagination as a model, we show that regulation of folded gastrulation expression by the Fork head transcription factor is required for apicomedial accumulation of Rho kinase and non-muscle myosin II, which coordinate apical constriction. We demonstrate that neither loss of spatially coordinated apical constriction nor its complete blockage prevent internalization and tube formation, although such manipulations affect the geometry of invagination. When apical constriction is disrupted, compressing force generated by a tissue-level myosin cable contributes to SG invagination. We demonstrate that fully elongated polarized SGs can form outside the embryo, suggesting that tube formation and elongation are intrinsic properties of the SG.
Keywords: D. melanogaster; Drosophila; apical constriction; cell biology; developmental biology; fog; fork head; myosin; salivary gland; stem cells.
Conflict of interest statement
The authors declare that no competing interests exist.
Figures




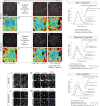

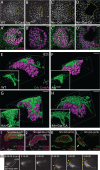


Similar articles
-
Regulation of apical constriction via microtubule- and Rab11-dependent apical transport during tissue invagination.Mol Biol Cell. 2021 May 1;32(10):1033-1047. doi: 10.1091/mbc.E21-01-0021. Epub 2021 Mar 31. Mol Biol Cell. 2021. PMID: 33788621 Free PMC article.
-
A three-dimensional vertex model forDrosophilasalivary gland invagination.Phys Biol. 2021 May 28;18(4). doi: 10.1088/1478-3975/abfa69. Phys Biol. 2021. PMID: 33882465
-
Downregulation of basal myosin-II is required for cell shape changes and tissue invagination.EMBO J. 2018 Dec 3;37(23):e100170. doi: 10.15252/embj.2018100170. Epub 2018 Nov 15. EMBO J. 2018. PMID: 30442834 Free PMC article.
-
Genetic regulation of salivary gland development in Drosophila melanogaster.Front Oral Biol. 2010;14:32-47. doi: 10.1159/000313706. Epub 2010 Apr 20. Front Oral Biol. 2010. PMID: 20428010 Review.
-
Controlling cell shape changes during salivary gland tube formation in Drosophila.Semin Cell Dev Biol. 2014 Jul;31:74-81. doi: 10.1016/j.semcdb.2014.03.020. Epub 2014 Mar 29. Semin Cell Dev Biol. 2014. PMID: 24685610 Review.
Cited by
-
Fog signaling has diverse roles in epithelial morphogenesis in insects.Elife. 2019 Oct 1;8:e47346. doi: 10.7554/eLife.47346. Elife. 2019. PMID: 31573513 Free PMC article.
-
The RhoGEF protein Plekhg5 regulates medioapical and junctional actomyosin dynamics of apical constriction during Xenopus gastrulation.Mol Biol Cell. 2023 Jun 1;34(7):ar64. doi: 10.1091/mbc.E22-09-0411. Epub 2023 Apr 12. Mol Biol Cell. 2023. PMID: 37043306 Free PMC article.
-
Template-based mapping of dynamic motifs in tissue morphogenesis.PLoS Comput Biol. 2020 Aug 21;16(8):e1008049. doi: 10.1371/journal.pcbi.1008049. eCollection 2020 Aug. PLoS Comput Biol. 2020. PMID: 32822341 Free PMC article.
-
Two-step regulation of trachealess ensures tight coupling of cell fate with morphogenesis in the Drosophila trachea.Elife. 2019 Aug 23;8:e45145. doi: 10.7554/eLife.45145. Elife. 2019. PMID: 31439126 Free PMC article.
-
Epidermal growth factor receptor signaling protects epithelia from morphogenetic instability and tissue damage in Drosophila.Development. 2023 Mar 1;150(5):dev201231. doi: 10.1242/dev.201231. Epub 2023 Mar 9. Development. 2023. PMID: 36897356 Free PMC article.
References
-
- Afshar K, Stuart B, Wasserman SA. Functional analysis of the Drosophila diaphanous FH protein in early embryonic development. Development. 2000;127:1887–1897. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

